An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- HCA Healthc J Med
- v.1(2); 2020
- PMC10324782


Introduction to Research Statistical Analysis: An Overview of the Basics
Christian vandever.
1 HCA Healthcare Graduate Medical Education
Description
This article covers many statistical ideas essential to research statistical analysis. Sample size is explained through the concepts of statistical significance level and power. Variable types and definitions are included to clarify necessities for how the analysis will be interpreted. Categorical and quantitative variable types are defined, as well as response and predictor variables. Statistical tests described include t-tests, ANOVA and chi-square tests. Multiple regression is also explored for both logistic and linear regression. Finally, the most common statistics produced by these methods are explored.
Introduction
Statistical analysis is necessary for any research project seeking to make quantitative conclusions. The following is a primer for research-based statistical analysis. It is intended to be a high-level overview of appropriate statistical testing, while not diving too deep into any specific methodology. Some of the information is more applicable to retrospective projects, where analysis is performed on data that has already been collected, but most of it will be suitable to any type of research. This primer will help the reader understand research results in coordination with a statistician, not to perform the actual analysis. Analysis is commonly performed using statistical programming software such as R, SAS or SPSS. These allow for analysis to be replicated while minimizing the risk for an error. Resources are listed later for those working on analysis without a statistician.
After coming up with a hypothesis for a study, including any variables to be used, one of the first steps is to think about the patient population to apply the question. Results are only relevant to the population that the underlying data represents. Since it is impractical to include everyone with a certain condition, a subset of the population of interest should be taken. This subset should be large enough to have power, which means there is enough data to deliver significant results and accurately reflect the study’s population.
The first statistics of interest are related to significance level and power, alpha and beta. Alpha (α) is the significance level and probability of a type I error, the rejection of the null hypothesis when it is true. The null hypothesis is generally that there is no difference between the groups compared. A type I error is also known as a false positive. An example would be an analysis that finds one medication statistically better than another, when in reality there is no difference in efficacy between the two. Beta (β) is the probability of a type II error, the failure to reject the null hypothesis when it is actually false. A type II error is also known as a false negative. This occurs when the analysis finds there is no difference in two medications when in reality one works better than the other. Power is defined as 1-β and should be calculated prior to running any sort of statistical testing. Ideally, alpha should be as small as possible while power should be as large as possible. Power generally increases with a larger sample size, but so does cost and the effect of any bias in the study design. Additionally, as the sample size gets bigger, the chance for a statistically significant result goes up even though these results can be small differences that do not matter practically. Power calculators include the magnitude of the effect in order to combat the potential for exaggeration and only give significant results that have an actual impact. The calculators take inputs like the mean, effect size and desired power, and output the required minimum sample size for analysis. Effect size is calculated using statistical information on the variables of interest. If that information is not available, most tests have commonly used values for small, medium or large effect sizes.
When the desired patient population is decided, the next step is to define the variables previously chosen to be included. Variables come in different types that determine which statistical methods are appropriate and useful. One way variables can be split is into categorical and quantitative variables. ( Table 1 ) Categorical variables place patients into groups, such as gender, race and smoking status. Quantitative variables measure or count some quantity of interest. Common quantitative variables in research include age and weight. An important note is that there can often be a choice for whether to treat a variable as quantitative or categorical. For example, in a study looking at body mass index (BMI), BMI could be defined as a quantitative variable or as a categorical variable, with each patient’s BMI listed as a category (underweight, normal, overweight, and obese) rather than the discrete value. The decision whether a variable is quantitative or categorical will affect what conclusions can be made when interpreting results from statistical tests. Keep in mind that since quantitative variables are treated on a continuous scale it would be inappropriate to transform a variable like which medication was given into a quantitative variable with values 1, 2 and 3.
Categorical vs. Quantitative Variables
Both of these types of variables can also be split into response and predictor variables. ( Table 2 ) Predictor variables are explanatory, or independent, variables that help explain changes in a response variable. Conversely, response variables are outcome, or dependent, variables whose changes can be partially explained by the predictor variables.
Response vs. Predictor Variables
Choosing the correct statistical test depends on the types of variables defined and the question being answered. The appropriate test is determined by the variables being compared. Some common statistical tests include t-tests, ANOVA and chi-square tests.
T-tests compare whether there are differences in a quantitative variable between two values of a categorical variable. For example, a t-test could be useful to compare the length of stay for knee replacement surgery patients between those that took apixaban and those that took rivaroxaban. A t-test could examine whether there is a statistically significant difference in the length of stay between the two groups. The t-test will output a p-value, a number between zero and one, which represents the probability that the two groups could be as different as they are in the data, if they were actually the same. A value closer to zero suggests that the difference, in this case for length of stay, is more statistically significant than a number closer to one. Prior to collecting the data, set a significance level, the previously defined alpha. Alpha is typically set at 0.05, but is commonly reduced in order to limit the chance of a type I error, or false positive. Going back to the example above, if alpha is set at 0.05 and the analysis gives a p-value of 0.039, then a statistically significant difference in length of stay is observed between apixaban and rivaroxaban patients. If the analysis gives a p-value of 0.91, then there was no statistical evidence of a difference in length of stay between the two medications. Other statistical summaries or methods examine how big of a difference that might be. These other summaries are known as post-hoc analysis since they are performed after the original test to provide additional context to the results.
Analysis of variance, or ANOVA, tests can observe mean differences in a quantitative variable between values of a categorical variable, typically with three or more values to distinguish from a t-test. ANOVA could add patients given dabigatran to the previous population and evaluate whether the length of stay was significantly different across the three medications. If the p-value is lower than the designated significance level then the hypothesis that length of stay was the same across the three medications is rejected. Summaries and post-hoc tests also could be performed to look at the differences between length of stay and which individual medications may have observed statistically significant differences in length of stay from the other medications. A chi-square test examines the association between two categorical variables. An example would be to consider whether the rate of having a post-operative bleed is the same across patients provided with apixaban, rivaroxaban and dabigatran. A chi-square test can compute a p-value determining whether the bleeding rates were significantly different or not. Post-hoc tests could then give the bleeding rate for each medication, as well as a breakdown as to which specific medications may have a significantly different bleeding rate from each other.
A slightly more advanced way of examining a question can come through multiple regression. Regression allows more predictor variables to be analyzed and can act as a control when looking at associations between variables. Common control variables are age, sex and any comorbidities likely to affect the outcome variable that are not closely related to the other explanatory variables. Control variables can be especially important in reducing the effect of bias in a retrospective population. Since retrospective data was not built with the research question in mind, it is important to eliminate threats to the validity of the analysis. Testing that controls for confounding variables, such as regression, is often more valuable with retrospective data because it can ease these concerns. The two main types of regression are linear and logistic. Linear regression is used to predict differences in a quantitative, continuous response variable, such as length of stay. Logistic regression predicts differences in a dichotomous, categorical response variable, such as 90-day readmission. So whether the outcome variable is categorical or quantitative, regression can be appropriate. An example for each of these types could be found in two similar cases. For both examples define the predictor variables as age, gender and anticoagulant usage. In the first, use the predictor variables in a linear regression to evaluate their individual effects on length of stay, a quantitative variable. For the second, use the same predictor variables in a logistic regression to evaluate their individual effects on whether the patient had a 90-day readmission, a dichotomous categorical variable. Analysis can compute a p-value for each included predictor variable to determine whether they are significantly associated. The statistical tests in this article generate an associated test statistic which determines the probability the results could be acquired given that there is no association between the compared variables. These results often come with coefficients which can give the degree of the association and the degree to which one variable changes with another. Most tests, including all listed in this article, also have confidence intervals, which give a range for the correlation with a specified level of confidence. Even if these tests do not give statistically significant results, the results are still important. Not reporting statistically insignificant findings creates a bias in research. Ideas can be repeated enough times that eventually statistically significant results are reached, even though there is no true significance. In some cases with very large sample sizes, p-values will almost always be significant. In this case the effect size is critical as even the smallest, meaningless differences can be found to be statistically significant.
These variables and tests are just some things to keep in mind before, during and after the analysis process in order to make sure that the statistical reports are supporting the questions being answered. The patient population, types of variables and statistical tests are all important things to consider in the process of statistical analysis. Any results are only as useful as the process used to obtain them. This primer can be used as a reference to help ensure appropriate statistical analysis.
Funding Statement
This research was supported (in whole or in part) by HCA Healthcare and/or an HCA Healthcare affiliated entity.
Conflicts of Interest
The author declares he has no conflicts of interest.
Christian Vandever is an employee of HCA Healthcare Graduate Medical Education, an organization affiliated with the journal’s publisher.
This research was supported (in whole or in part) by HCA Healthcare and/or an HCA Healthcare affiliated entity. The views expressed in this publication represent those of the author(s) and do not necessarily represent the official views of HCA Healthcare or any of its affiliated entities.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Statistics articles within Scientific Reports
Article 23 May 2024 | Open Access
Analysis of player speed and angle toward the ball in soccer
- Álvaro Novillo
- , Antonio Cordón-Carmona
- & Javier M. Buldú
Article 15 May 2024 | Open Access
Modified correlated measurement errors model for estimation of population mean utilizing auxiliary information
- Housila P. Singh
- & Neha Garg
Article 14 May 2024 | Open Access
Employing machine learning for enhanced abdominal fat prediction in cavitation post-treatment
- Doaa A. Abdel Hady
- , Omar M. Mabrouk
- & Tarek Abd El-Hafeez
Article 09 May 2024 | Open Access
Towards optimal model evaluation: enhancing active testing with actively improved estimators
- JooChul Lee
- , Likhitha Kolla
- & Jinbo Chen
Article 08 May 2024 | Open Access
Research on proactive defense and dynamic repair of complex networks considering cascading effects
- Zhuoying Shi
- , Ying Wang
- & Chaoqi Fu
Article 07 May 2024 | Open Access
A Markov network approach for reproducing purchase behaviours observed in convenience stores
- Dan Johansson
- , Hideki Takayasu
- & Misako Takayasu
Article 06 May 2024 | Open Access
A Bayesian spatio-temporal model of COVID-19 spread in England
- Xueqing Yin
- , John M. Aiken
- & Jonathan L. Bamber
Max-mixed EWMA control chart for joint monitoring of mean and variance: an application to yogurt packing process
- Seher Malik
- , Muhammad Hanif
- & Jumanah Ahmed Darwish
Causal impact evaluation of occupational safety policies on firms’ default using machine learning uplift modelling
- Berardino Barile
- , Marco Forti
- & Angelo Castaldo
Article 04 May 2024 | Open Access
Estimating neutrosophic finite median employing robust measures of the auxiliary variable
- Saadia Masood
- , Bareera Ibrar
- & Zabihullah Movaheedi
Article 01 May 2024 | Open Access
Zika emergence, persistence, and transmission rate in Colombia: a nationwide application of a space-time Markov switching model
- Laís Picinini Freitas
- , Dirk Douwes-Schultz
- & Kate Zinszer
Article 29 April 2024 | Open Access
Exploring drivers of overnight stays and same-day visits in the tourism sector
- Francesco Scotti
- , Andrea Flori
- & Giovanni Azzone
A support vector machine based drought index for regional drought analysis
- Mohammed A Alshahrani
- , Muhammad Laiq
- & Muhammad Nabi
Article 25 April 2024 | Open Access
Joint Bayesian estimation of cell dependence and gene associations in spatially resolved transcriptomic data
- Arhit Chakrabarti
- & Bani K. Mallick
Estimating SARS-CoV-2 infection probabilities with serological data and a Bayesian mixture model
- Benjamin Glemain
- , Xavier de Lamballerie
- & Fabrice Carrat
Article 24 April 2024 | Open Access
Applications of nature-inspired metaheuristic algorithms for tackling optimization problems across disciplines
- Elvis Han Cui
- , Zizhao Zhang
- & Weng Kee Wong
Article 23 April 2024 | Open Access
Variable parameters memory-type control charts for simultaneous monitoring of the mean and variability of multivariate multiple linear regression profiles
- Hamed Sabahno
- & Marie Eriksson
Article 22 April 2024 | Open Access
Modeling health and well-being measures using ZIP code spatial neighborhood patterns
- , Michael LaValley
- & Shariq Mohammed
Article 20 April 2024 | Open Access
Sequence based model using deep neural network and hybrid features for identification of 5-hydroxymethylcytosine modification
- Salman Khan
- , Islam Uddin
- & Dost Muhammad Khan
Article 19 April 2024 | Open Access
Identification of CT radiomic features robust to acquisition and segmentation variations for improved prediction of radiotherapy-treated lung cancer patient recurrence
- Thomas Louis
- , François Lucia
- & Roland Hustinx
Explainable prediction of node labels in multilayer networks: a case study of turnover prediction in organizations
- László Gadár
- & János Abonyi
Article 18 April 2024 | Open Access
The quasi-xgamma frailty model with survival analysis under heterogeneity problem, validation testing, and risk analysis for emergency care data
- Hamami Loubna
- , Hafida Goual
- & Haitham M. Yousof
Memory type Bayesian adaptive max-EWMA control chart for weibull processes
- Abdullah A. Zaagan
- , Imad Khan
- & Bakhtiyar Ahmad
Article 17 April 2024 | Open Access
Improved data quality and statistical power of trial-level event-related potentials with Bayesian random-shift Gaussian processes
- Dustin Pluta
- , Beniamino Hadj-Amar
- & Marina Vannucci
Article 16 April 2024 | Open Access
Comparison and evaluation of overcoring and hydraulic fracturing stress measurements
- , Meifeng Cai
- & Mostafa Gorjian
Predictors of divorce and duration of marriage among first marriage women in Dejne administrative town
- Nigusie Gashaye Shita
- & Liknaw Bewket Zeleke
Article 12 April 2024 | Open Access
Determinants of multimodal fake review generation in China’s E-commerce platforms
- Chunnian Liu
- & Lan Yi
Article 11 April 2024 | Open Access
New ridge parameter estimators for the quasi-Poisson ridge regression model
- Aamir Shahzad
- , Muhammad Amin
- & Muhammad Faisal
A bicoherence approach to analyze multi-dimensional cross-frequency coupling in EEG/MEG data
- Alessio Basti
- , Guido Nolte
- & Laura Marzetti
Article 10 April 2024 | Open Access
Response times are affected by mispredictions in a stochastic game
- Paulo Roberto Cabral-Passos
- , Antonio Galves
- & Claudia D. Vargas
The effect of city reputation on Chinese corporate risk-taking
- & Haifeng Jiang
Article 06 April 2024 | Open Access
Improvement in variance estimation using transformed auxiliary variable under simple random sampling
- , Syed Muhammad Asim
- & Soofia Iftikhar
Article 28 March 2024 | Open Access
Fatty liver classification via risk controlled neural networks trained on grouped ultrasound image data
- Tso-Jung Yen
- , Chih-Ting Yang
- & Hsin-Chou Yang
Article 27 March 2024 | Open Access
A new unit distribution: properties, estimation, and regression analysis
- Kadir Karakaya
- , C. S. Rajitha
- & Ahmed M. Gemeay
Article 26 March 2024 | Open Access
GeneAI 3.0: powerful, novel, generalized hybrid and ensemble deep learning frameworks for miRNA species classification of stationary patterns from nucleotides
- Jaskaran Singh
- , Narendra N. Khanna
- & Jasjit S. Suri
On topological indices and entropy measures of beryllonitrene network via logarithmic regression model
- , Muhammad Kamran Siddiqui
- & Fikre Bogale Petros
Article 22 March 2024 | Open Access
Measuring the similarity of charts in graphical statistics
- Krzysztof Górnisiewicz
- , Zbigniew Palka
- & Waldemar Ratajczak
Article 21 March 2024 | Open Access
Risk prediction and interaction analysis using polygenic risk score of type 2 diabetes in a Korean population
- Minsun Song
- , Soo Heon Kwak
- & Jihyun Kim
A longitudinal causal graph analysis investigating modifiable risk factors and obesity in a European cohort of children and adolescents
- Ronja Foraita
- , Janine Witte
- & Vanessa Didelez
Article 19 March 2024 | Open Access
A novel group decision making method based on CoCoSo and interval-valued Q-rung orthopair fuzzy sets
- , Hongwu Qin
- & Xiuqin Ma
Impact of using virtual avatars in educational videos on user experience
- Ruyuan Zhang
- & Qun Wu
A generalisation of the method of regression calibration and comparison with Bayesian and frequentist model averaging methods
- Mark P. Little
- , Nobuyuki Hamada
- & Lydia B. Zablotska
Article 18 March 2024 | Open Access
Monitoring gamma type-I censored data using an exponentially weighted moving average control chart based on deep learning networks
- Pei-Hsi Lee
- & Shih-Lung Liao
Article 15 March 2024 | Open Access
Statistical detection of selfish mining in proof-of-work blockchain systems
- Sheng-Nan Li
- , Carlo Campajola
- & Claudio J. Tessone
Article 13 March 2024 | Open Access
Evaluation metrics and statistical tests for machine learning
- Oona Rainio
- , Jarmo Teuho
- & Riku Klén
PARSEG: a computationally efficient approach for statistical validation of botanical seeds’ images
- Luca Frigau
- , Claudio Conversano
- & Jaromír Antoch
Application of analysis of variance to determine important features of signals for diagnostic classifiers of displacement pumps
- Jarosław Konieczny
- , Waldemar Łatas
- & Jerzy Stojek
Article 12 March 2024 | Open Access
Prediction and detection of side effects severity following COVID-19 and influenza vaccinations: utilizing smartwatches and smartphones
- , Margaret L. Brandeau
- & Dan Yamin
Article 08 March 2024 | Open Access
Evaluating the lifetime performance index of omega distribution based on progressive type-II censored samples
- N. M. Kilany
- & Lobna H. El-Refai
Article 07 March 2024 | Open Access
Development of risk models of incident hypertension using machine learning on the HUNT study data
- Filip Emil Schjerven
- , Emma Maria Lovisa Ingeström
- & Frank Lindseth
Browse broader subjects
- Mathematics and computing
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Reporting statistical methods and outcome of statistical analyses in research articles
- Published: 15 June 2020
- Volume 72 , pages 481–485, ( 2020 )
Cite this article

- Mariusz Cichoń 1
17k Accesses
13 Citations
1 Altmetric
Explore all metrics
Avoid common mistakes on your manuscript.
Introduction
Statistical methods constitute a powerful tool in modern life sciences. This tool is primarily used to disentangle whether the observed differences, relationships or congruencies are meaningful or may just occur by chance. Thus, statistical inference is an unavoidable part of scientific work. The knowledge of statistics is usually quite limited among researchers representing the field of life sciences, particularly when it comes to constraints imposed on the use of statistical tools and possible interpretations. A common mistake is that researchers take for granted the ability to perform a valid statistical analysis. However, at the stage of data analysis, it may turn out that the gathered data cannot be analysed with any known statistical tools or that there are critical flaws in the interpretation of the results due to violations of basic assumptions of statistical methods. A common mistake made by authors is to thoughtlessly copy the choice of the statistical tests from other authors analysing similar data. This strategy, although sometimes correct, may lead to an incorrect choice of statistical tools and incorrect interpretations. Here, I aim to give some advice on how to choose suitable statistical methods and how to present the results of statistical analyses.
Important limits in the use of statistics
Statistical tools face a number of constraints. Constraints should already be considered at the stage of planning the research, as mistakes made at this stage may make statistical analyses impossible. Therefore, careful planning of sampling is critical for future success in data analyses. The most important is ensuring that the general population is sampled randomly and independently, and that the experimental design corresponds to the aims of the research. Planning a control group/groups is of particular importance. Without a suitable control group, any further inference may not be possible. Parametric tests are stronger (it is easier to reject a null hypothesis), so they should always be preferred, but such methods can be used only when the data are drawn from a general population with normal distribution. For methods based on analysis of variance (ANOVA), residuals should come from a general population with normal distribution, and in this case there is an additional important assumption of homogeneity of variance. Inferences made from analyses violating these assumptions may be incorrect.
Statistical inference
Statistical inference is asymmetrical. Scientific discovery is based on rejecting null hypotheses, so interpreting non-significant results should be taken with special care. We never know for sure why we fail to reject the null hypothesis. It may indeed be true, but it is also possible that our sample size was too small or variance too large to capture the differences or relationships. We also may fail just by chance. Assuming a significance level of p = 0.05 means that we run the risk of rejecting a null hypothesis in 5% of such analyses. Thus, interpretation of non-significant results should always be accompanied by the so-called power analysis, which shows the strength of our inference.
Experimental design and data analyses
The experimental design is a critical part of study planning. The design must correspond to the aims of the study presented in the Introduction section. In turn, the statistical methods must be suited to the experimental design so that the data analyses will enable the questions stated in the Introduction to be answered. In general, simple experimental designs allow the use of simple methods like t-tests, simple correlations, etc., while more complicated designs (multifactor designs) require more advanced methods (see, Fig. 1 ). Data coming from more advanced designs usually cannot be analysed with simple methods. Therefore, multifactor designs cannot be followed by a simple t-test or even with one-way ANOVA, as factors may not act independently, and in such a case the interpretation of the results of one-way ANOVA may be incorrect. Here, it is particularly important that one may be interested in a concerted action of factors (interaction) or an action of a given factor while controlling for other factors (independent action of a factor). But even with one factor design with more than two levels, one cannot use just a simple t-test with multiple comparisons between groups. In such a case, one-way ANOVA should be performed followed by a post hoc test. The post hoc test can be done only if ANOVA rejects the null hypothesis. There is no point in using the post hoc test if the factors have only two levels (groups). In this case, the differences are already clear after ANOVA.
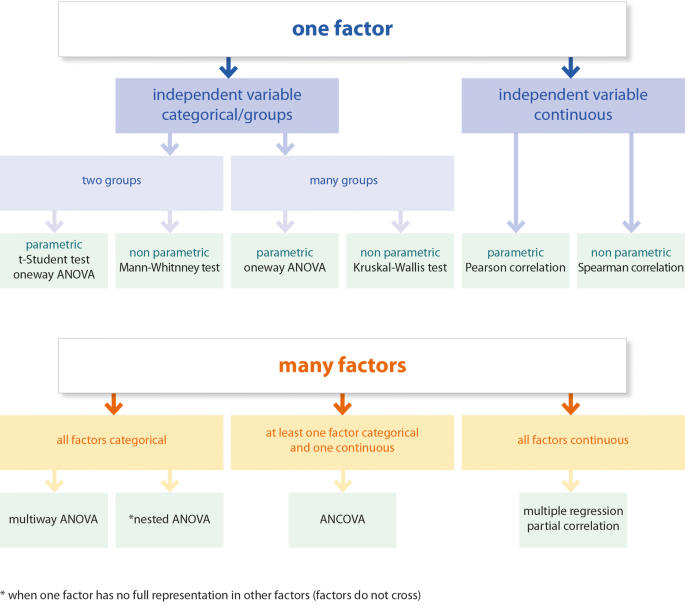
Test selection chart
Description of statistical methods in the Materials and methods section
It is in the author’s interest to provide the reader with all necessary information to judge whether the statistical tools used in the paper are the most suitable to answer the scientific question and are suited to the data structure. In the Materials and methods section, the experimental design must be described in detail, so that the reader may easily understand how the study was performed and later why such specific statistical methods were chosen. It must be clear whether the study is planned to test the relationships or differences between groups. Here, the reader should already understand the data structure, what the dependent variable is, what the factors are, and should be able to determine, even without being directly informed, whether the factors are categorical or continuous, and whether they are fixed or random. The sample size used in the analysis should be clearly stated. Sometimes sample sizes used in analyses are smaller than the original. This can happen for various reasons, for example if one fails to perform some measurements, and in such a case, the authors must clearly explain why the original sample size differs from the one used in the analyses. There must be a very good reason to omit existing data points from the analyses. Removing the so-called outliers should be an exception rather than the rule.
A description of the statistical methods should come at the end of the Materials and methods section. Here, we start by introducing the statistical techniques used to test predictions formulated in the Introduction. We describe in detail the structure of the statistical model (defining the dependent variable, the independent variables—factors, interactions if present, character of the factors—fixed or random). The variables should be defined as categorical or continuous. In the case of more advanced models, information on the methods of effects estimation or degrees of freedom should be provided. Unless there are good reasons, interactions should always be tested, even if the study is not aimed at testing an interaction. If the interaction is not the main aim of the study, non-significant interactions should be dropped from the model and new analyses without interactions should be carried out and such results reported. If the interaction appears to be significant, one cannot remove it from the model even if the interaction is not the main aim of the study. In such a case, only the interaction can be interpreted, while the interpretation of the main effects is not allowed. The author should clearly describe how the interactions will be dealt with. One may also consider using a model selection procedure which should also be clearly described.
The authors should reassure the reader that the assumptions of the selected statistical technique are fully met. It must be described how the normality of data distribution and homogeneity of variance was checked and whether these assumptions have been met. When performing data transformation, one needs to explain how it was done and whether the transformation helped to fulfil the assumptions of the parametric tests. If these assumptions are not fulfilled, one may apply non-parametric tests. It must be clearly stated why non-parametric tests are performed. Post hoc tests can be performed only when the ANOVA/Kruskal–Wallis test shows significant effects. These tests are valid for the main effects only when the interaction is not included in the model. These tests are also applicable for significant interactions. There are a number of different post hoc tests, so the selected test must be introduced in the materials and methods section.
The significance level is often mentioned in the materials and methods section. There is common consensus among researchers in life sciences for a significance level set at p = 0.05, so it is not strictly necessary to report this conventional level unless the authors always give the I type error (p-value) throughout the paper. If the author sets the significance level at a lower value, which could be the case, for example, in medical sciences, the reader must be informed about the use of a more conservative level. If the significance level is not reported, the reader will assume p = 0.05. In general, it does not matter which statistical software was used for the analyses. However, the outcome may differ slightly between different software, even if exactly the same model is set. Thus, it may be a good practice to report the name of the software at the end of the subsection describing the statistical methods. If the original code of the model analysed is provided, it would be sensible to inform the reader of the specific software and version that was used.
Presentation of the outcome in the Results section
Only the data and the analyses needed to test the hypotheses and predictions stated in the Introduction and those important for discussion should be placed in the Results section. All other outcome might be provided as supplementary materials. Some descriptive statistics are often reported in the Results section, such as means, standard errors (SE), standard deviation (SD), confidence interval (CI). It is of critical importance that these estimates can only be provided if the described data are drawn from a general population with normal distribution; otherwise median values with quartiles should be provided. A common mistake is to provide the results of non-parametric tests with parametric estimates. If one cannot assume normal distribution, providing arithmetic mean with standard deviation is misleading, as they are estimates of normal distribution. I recommend using confidence intervals instead of SE or SD, as confidence intervals are more informative (non-overlapping intervals suggest the existence of potential differences).
Descriptive statistics can be calculated from raw data (measured values) or presented as estimates from the calculated models (values corrected for independent effects of other factors in the model). The issue whether estimates from models or statistics calculated from the raw data provided throughout the paper should be clearly stated in the Materials and methods section. It is not necessary to report the descriptive statistics in the text if it is already reported in the tables or can be easily determined from the graphs.
The Results section is a narrative text which tells the reader about all the findings and guides them to refer to tables and figures if present. Each table and figure should be referenced in the text at least once. It is in the author’s interest to provide the reader the outcome of the statistical tests in such a way that the correctness of the reported values can be assessed. The value of the appropriate statistics (e.g. F, t, H, U, z, r) must always be provided, along with the sample size (N; non-parametric tests) or degrees of freedom (df; parametric tests) and I type error (p-value). The p-value is an important information, as it tells the reader about confidence related to rejecting the null hypothesis. Thus one needs to provide an exact value of I type error. A common mistake is to provide information as an inequality (p < 0.05). There is an important difference for interpretation if p = 0.049 or p = 0.001.
The outcome of simple tests (comparing two groups, testing relationship between two variables) can easily be reported in the text, but in case of multivariate models, one may rather report the outcome in the form of a table in which all factors with their possible interactions are listed with their estimates, statistics and p-values. The results of post hoc tests, if performed, may be reported in the main text, but if one reports differences between many groups or an interaction, then presenting such results in the form of a table or graph could be more informative.
The main results are often presented graphically, particularly when the effects appear to be significant. The graphs should be constructed so that they correspond to the analyses. If the main interest of the study is in an interaction, then it should be depicted in the graph. One should not present interaction in the graph if it appeared to be non-significant. When presenting differences, the mean or median value should be visualised as a dot, circle or some other symbol with some measure of variability (quartiles if a non-parametric test was performed, and SD, SE or preferably confidence intervals in the case of parametric tests) as whiskers below and above the midpoint. The midpoints should not be linked with a line unless an interaction is presented or, more generally, if the line has some biological/logical meaning in the experimental design. Some authors present differences as bar graphs. When using bar graphs, the Y -axis must start from a zero value. If a bar graph is used to show differences between groups, some measure of variability (SD, SE, CI) must also be provided, as whiskers, for example. Graphs may present the outcome of post hoc tests in the form of letters placed above the midpoint or whiskers, with the same letter indicating lack of differences and different letters signalling pairwise differences. The significant differences can also be denoted as asterisks or, preferably, p-values placed above the horizontal line linking the groups. All this must be explained in the figure caption. Relationships should be presented in the form of a scatterplot. This could be accompanied by a regression line, but only if the relationship is statistically significant. The regression line is necessary if one is interested in describing a functional relationship between two variables. If one is interested in correlation between variables, the regression line is not necessary, but could be placed in order to visualise the relationship. In this case, it must be explained in the figure caption. If regression is of interest, then providing an equation of this regression is necessary in the figure caption. Remember that graphs serve to represent the analyses performed, so if the analyses were carried out on the transformed data, the graphs should also present transformed data. In general, the tables and figure captions must be self-explanatory, so that the reader is able to understand the table/figure content without reading the main text. The table caption should be written in such a way that it is possible to understand the statistical analysis from which the results are presented.
Guidelines for the Materials and methods section:
Provide detailed description of the experimental design so that the statistical techniques will be understandable for the reader.
Make sure that factors and groups within factors are clearly introduced.
Describe all statistical techniques applied in the study and provide justification for each test (both parametric and non-parametric methods).
If parametric tests are used, describe how the normality of data distribution and homogeneity of variance (in the case of analysis of variance) was checked and state clearly that these important assumptions for parametric tests are met.
Give a rationale for using non-parametric tests.
If data transformation was applied, provide details of how this transformation was performed and state clearly that this helped to achieve normal distribution/homogeneity of variance.
In the case of multivariate analyses, describe the statistical model in detail and explain what you did with interactions.
If post hoc tests are used, clearly state which tests you use.
Specify the type of software and its version if you think it is important.
Guidelines for presentation of the outcome of statistical analyses in the Results section:
Make sure you report appropriate descriptive statistics—means, standard errors (SE), standard deviation (SD), confidence intervals (CI), etc. in case of parametric tests or median values with quartiles in case of non-parametric tests.
Provide appropriate statistics for your test (t value for t-test, F for ANOVA, H for Kruskal–Wallis test, U for Mann–Whitney test, χ 2 for chi square test, or r for correlation) along with the sample size (non-parametric tests) or degrees of freedom (df; parametric tests).
t 23 = 3.45 (the number in the subscript denotes degree of freedom, meaning the sample size of the first group minus 1 plus the sample size of the second group minus 1 for the test with independent groups, or number of pairs in paired t-test minus 1).
F 1,23 = 6.04 (first number in the subscript denotes degrees of freedom for explained variance—number of groups within factor minus 1, second number denotes degree of freedom for unexplained variance—residual variance). F-statistics should be provided separately for all factors and interactions (only if interactions are present in the model).
H = 13.8, N 1 = 15, N 2 = 18, N 3 = 12 (N 1, N 2, N 3 are sample sizes for groups compared).
U = 50, N 1 = 20, N 2 = 19 for Mann–Whitney test (N 1 and N 2 are sample sizes for groups).
χ 2 = 3.14 df = 1 (here meaning e.g. 2 × 2 contingency table).
r = 0.78, N = 32 or df = 30 (df = N − 2).
Provide exact p-values (e.g. p = 0.03), rather than standard inequality (p ≤ 0.05)
If the results of statistical analysis are presented in the form of a table, make sure the statistical model is accurately described so that the reader will understand the context of the table without referring to the text. Please ensure that the table is cited in the text.
The figure caption should include all information necessary to understand what is seen in the figure. Describe what is denoted by a bar, symbols, whiskers (mean/median, SD, SE, CI/quartiles). If you present transformed data, inform the reader about the transformation you applied. If you present the results of a post hoc test on the graph, please note what test was used and how you denote the significant differences. If you present a regression line on the scatter plot, give information as to whether you provide the line to visualise the relationship or you are indeed interested in regression, and in the latter case, give the equation for this regression line.
Further reading in statistics:
Sokal and Rolf. 2011. Biometry. Freeman.
Zar. 2010. Biostatistical analyses. Prentice Hall.
McDonald, J.H. 2014. Handbook of biological statistics. Sparky House Publishing, Baltimore, Maryland.
Quinn and Keough. 2002. Experimental design and data analysis for biologists. Cambridge University Press.
Author information
Authors and affiliations.
Institute of Environmental Sciences, Jagiellonian University, Gronostajowa 7, 30-376, Kraków, Poland
Mariusz Cichoń
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mariusz Cichoń .
Rights and permissions
Reprints and permissions
About this article
Cichoń, M. Reporting statistical methods and outcome of statistical analyses in research articles. Pharmacol. Rep 72 , 481–485 (2020). https://doi.org/10.1007/s43440-020-00110-5
Download citation
Published : 15 June 2020
Issue Date : June 2020
DOI : https://doi.org/10.1007/s43440-020-00110-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Find a journal
- Publish with us
- Track your research
Have a thesis expert improve your writing
Check your thesis for plagiarism in 10 minutes, generate your apa citations for free.
- Knowledge Base
The Beginner's Guide to Statistical Analysis | 5 Steps & Examples
Statistical analysis means investigating trends, patterns, and relationships using quantitative data . It is an important research tool used by scientists, governments, businesses, and other organisations.
To draw valid conclusions, statistical analysis requires careful planning from the very start of the research process . You need to specify your hypotheses and make decisions about your research design, sample size, and sampling procedure.
After collecting data from your sample, you can organise and summarise the data using descriptive statistics . Then, you can use inferential statistics to formally test hypotheses and make estimates about the population. Finally, you can interpret and generalise your findings.
This article is a practical introduction to statistical analysis for students and researchers. We’ll walk you through the steps using two research examples. The first investigates a potential cause-and-effect relationship, while the second investigates a potential correlation between variables.
Table of contents
Step 1: write your hypotheses and plan your research design, step 2: collect data from a sample, step 3: summarise your data with descriptive statistics, step 4: test hypotheses or make estimates with inferential statistics, step 5: interpret your results, frequently asked questions about statistics.
To collect valid data for statistical analysis, you first need to specify your hypotheses and plan out your research design.
Writing statistical hypotheses
The goal of research is often to investigate a relationship between variables within a population . You start with a prediction, and use statistical analysis to test that prediction.
A statistical hypothesis is a formal way of writing a prediction about a population. Every research prediction is rephrased into null and alternative hypotheses that can be tested using sample data.
While the null hypothesis always predicts no effect or no relationship between variables, the alternative hypothesis states your research prediction of an effect or relationship.
- Null hypothesis: A 5-minute meditation exercise will have no effect on math test scores in teenagers.
- Alternative hypothesis: A 5-minute meditation exercise will improve math test scores in teenagers.
- Null hypothesis: Parental income and GPA have no relationship with each other in college students.
- Alternative hypothesis: Parental income and GPA are positively correlated in college students.
Planning your research design
A research design is your overall strategy for data collection and analysis. It determines the statistical tests you can use to test your hypothesis later on.
First, decide whether your research will use a descriptive, correlational, or experimental design. Experiments directly influence variables, whereas descriptive and correlational studies only measure variables.
- In an experimental design , you can assess a cause-and-effect relationship (e.g., the effect of meditation on test scores) using statistical tests of comparison or regression.
- In a correlational design , you can explore relationships between variables (e.g., parental income and GPA) without any assumption of causality using correlation coefficients and significance tests.
- In a descriptive design , you can study the characteristics of a population or phenomenon (e.g., the prevalence of anxiety in U.S. college students) using statistical tests to draw inferences from sample data.
Your research design also concerns whether you’ll compare participants at the group level or individual level, or both.
- In a between-subjects design , you compare the group-level outcomes of participants who have been exposed to different treatments (e.g., those who performed a meditation exercise vs those who didn’t).
- In a within-subjects design , you compare repeated measures from participants who have participated in all treatments of a study (e.g., scores from before and after performing a meditation exercise).
- In a mixed (factorial) design , one variable is altered between subjects and another is altered within subjects (e.g., pretest and posttest scores from participants who either did or didn’t do a meditation exercise).
- Experimental
- Correlational
First, you’ll take baseline test scores from participants. Then, your participants will undergo a 5-minute meditation exercise. Finally, you’ll record participants’ scores from a second math test.
In this experiment, the independent variable is the 5-minute meditation exercise, and the dependent variable is the math test score from before and after the intervention. Example: Correlational research design In a correlational study, you test whether there is a relationship between parental income and GPA in graduating college students. To collect your data, you will ask participants to fill in a survey and self-report their parents’ incomes and their own GPA.
Measuring variables
When planning a research design, you should operationalise your variables and decide exactly how you will measure them.
For statistical analysis, it’s important to consider the level of measurement of your variables, which tells you what kind of data they contain:
- Categorical data represents groupings. These may be nominal (e.g., gender) or ordinal (e.g. level of language ability).
- Quantitative data represents amounts. These may be on an interval scale (e.g. test score) or a ratio scale (e.g. age).
Many variables can be measured at different levels of precision. For example, age data can be quantitative (8 years old) or categorical (young). If a variable is coded numerically (e.g., level of agreement from 1–5), it doesn’t automatically mean that it’s quantitative instead of categorical.
Identifying the measurement level is important for choosing appropriate statistics and hypothesis tests. For example, you can calculate a mean score with quantitative data, but not with categorical data.
In a research study, along with measures of your variables of interest, you’ll often collect data on relevant participant characteristics.
In most cases, it’s too difficult or expensive to collect data from every member of the population you’re interested in studying. Instead, you’ll collect data from a sample.
Statistical analysis allows you to apply your findings beyond your own sample as long as you use appropriate sampling procedures . You should aim for a sample that is representative of the population.
Sampling for statistical analysis
There are two main approaches to selecting a sample.
- Probability sampling: every member of the population has a chance of being selected for the study through random selection.
- Non-probability sampling: some members of the population are more likely than others to be selected for the study because of criteria such as convenience or voluntary self-selection.
In theory, for highly generalisable findings, you should use a probability sampling method. Random selection reduces sampling bias and ensures that data from your sample is actually typical of the population. Parametric tests can be used to make strong statistical inferences when data are collected using probability sampling.
But in practice, it’s rarely possible to gather the ideal sample. While non-probability samples are more likely to be biased, they are much easier to recruit and collect data from. Non-parametric tests are more appropriate for non-probability samples, but they result in weaker inferences about the population.
If you want to use parametric tests for non-probability samples, you have to make the case that:
- your sample is representative of the population you’re generalising your findings to.
- your sample lacks systematic bias.
Keep in mind that external validity means that you can only generalise your conclusions to others who share the characteristics of your sample. For instance, results from Western, Educated, Industrialised, Rich and Democratic samples (e.g., college students in the US) aren’t automatically applicable to all non-WEIRD populations.
If you apply parametric tests to data from non-probability samples, be sure to elaborate on the limitations of how far your results can be generalised in your discussion section .
Create an appropriate sampling procedure
Based on the resources available for your research, decide on how you’ll recruit participants.
- Will you have resources to advertise your study widely, including outside of your university setting?
- Will you have the means to recruit a diverse sample that represents a broad population?
- Do you have time to contact and follow up with members of hard-to-reach groups?
Your participants are self-selected by their schools. Although you’re using a non-probability sample, you aim for a diverse and representative sample. Example: Sampling (correlational study) Your main population of interest is male college students in the US. Using social media advertising, you recruit senior-year male college students from a smaller subpopulation: seven universities in the Boston area.
Calculate sufficient sample size
Before recruiting participants, decide on your sample size either by looking at other studies in your field or using statistics. A sample that’s too small may be unrepresentative of the sample, while a sample that’s too large will be more costly than necessary.
There are many sample size calculators online. Different formulas are used depending on whether you have subgroups or how rigorous your study should be (e.g., in clinical research). As a rule of thumb, a minimum of 30 units or more per subgroup is necessary.
To use these calculators, you have to understand and input these key components:
- Significance level (alpha): the risk of rejecting a true null hypothesis that you are willing to take, usually set at 5%.
- Statistical power : the probability of your study detecting an effect of a certain size if there is one, usually 80% or higher.
- Expected effect size : a standardised indication of how large the expected result of your study will be, usually based on other similar studies.
- Population standard deviation: an estimate of the population parameter based on a previous study or a pilot study of your own.
Once you’ve collected all of your data, you can inspect them and calculate descriptive statistics that summarise them.
Inspect your data
There are various ways to inspect your data, including the following:
- Organising data from each variable in frequency distribution tables .
- Displaying data from a key variable in a bar chart to view the distribution of responses.
- Visualising the relationship between two variables using a scatter plot .
By visualising your data in tables and graphs, you can assess whether your data follow a skewed or normal distribution and whether there are any outliers or missing data.
A normal distribution means that your data are symmetrically distributed around a center where most values lie, with the values tapering off at the tail ends.
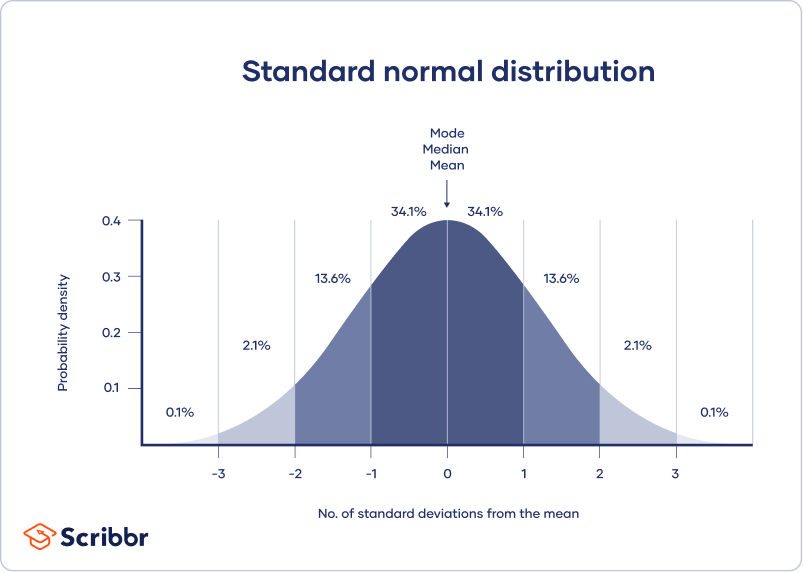
In contrast, a skewed distribution is asymmetric and has more values on one end than the other. The shape of the distribution is important to keep in mind because only some descriptive statistics should be used with skewed distributions.
Extreme outliers can also produce misleading statistics, so you may need a systematic approach to dealing with these values.
Calculate measures of central tendency
Measures of central tendency describe where most of the values in a data set lie. Three main measures of central tendency are often reported:
- Mode : the most popular response or value in the data set.
- Median : the value in the exact middle of the data set when ordered from low to high.
- Mean : the sum of all values divided by the number of values.
However, depending on the shape of the distribution and level of measurement, only one or two of these measures may be appropriate. For example, many demographic characteristics can only be described using the mode or proportions, while a variable like reaction time may not have a mode at all.
Calculate measures of variability
Measures of variability tell you how spread out the values in a data set are. Four main measures of variability are often reported:
- Range : the highest value minus the lowest value of the data set.
- Interquartile range : the range of the middle half of the data set.
- Standard deviation : the average distance between each value in your data set and the mean.
- Variance : the square of the standard deviation.
Once again, the shape of the distribution and level of measurement should guide your choice of variability statistics. The interquartile range is the best measure for skewed distributions, while standard deviation and variance provide the best information for normal distributions.
Using your table, you should check whether the units of the descriptive statistics are comparable for pretest and posttest scores. For example, are the variance levels similar across the groups? Are there any extreme values? If there are, you may need to identify and remove extreme outliers in your data set or transform your data before performing a statistical test.
From this table, we can see that the mean score increased after the meditation exercise, and the variances of the two scores are comparable. Next, we can perform a statistical test to find out if this improvement in test scores is statistically significant in the population. Example: Descriptive statistics (correlational study) After collecting data from 653 students, you tabulate descriptive statistics for annual parental income and GPA.
It’s important to check whether you have a broad range of data points. If you don’t, your data may be skewed towards some groups more than others (e.g., high academic achievers), and only limited inferences can be made about a relationship.
A number that describes a sample is called a statistic , while a number describing a population is called a parameter . Using inferential statistics , you can make conclusions about population parameters based on sample statistics.
Researchers often use two main methods (simultaneously) to make inferences in statistics.
- Estimation: calculating population parameters based on sample statistics.
- Hypothesis testing: a formal process for testing research predictions about the population using samples.
You can make two types of estimates of population parameters from sample statistics:
- A point estimate : a value that represents your best guess of the exact parameter.
- An interval estimate : a range of values that represent your best guess of where the parameter lies.
If your aim is to infer and report population characteristics from sample data, it’s best to use both point and interval estimates in your paper.
You can consider a sample statistic a point estimate for the population parameter when you have a representative sample (e.g., in a wide public opinion poll, the proportion of a sample that supports the current government is taken as the population proportion of government supporters).
There’s always error involved in estimation, so you should also provide a confidence interval as an interval estimate to show the variability around a point estimate.
A confidence interval uses the standard error and the z score from the standard normal distribution to convey where you’d generally expect to find the population parameter most of the time.
Hypothesis testing
Using data from a sample, you can test hypotheses about relationships between variables in the population. Hypothesis testing starts with the assumption that the null hypothesis is true in the population, and you use statistical tests to assess whether the null hypothesis can be rejected or not.
Statistical tests determine where your sample data would lie on an expected distribution of sample data if the null hypothesis were true. These tests give two main outputs:
- A test statistic tells you how much your data differs from the null hypothesis of the test.
- A p value tells you the likelihood of obtaining your results if the null hypothesis is actually true in the population.
Statistical tests come in three main varieties:
- Comparison tests assess group differences in outcomes.
- Regression tests assess cause-and-effect relationships between variables.
- Correlation tests assess relationships between variables without assuming causation.
Your choice of statistical test depends on your research questions, research design, sampling method, and data characteristics.

Parametric tests
Parametric tests make powerful inferences about the population based on sample data. But to use them, some assumptions must be met, and only some types of variables can be used. If your data violate these assumptions, you can perform appropriate data transformations or use alternative non-parametric tests instead.
A regression models the extent to which changes in a predictor variable results in changes in outcome variable(s).
- A simple linear regression includes one predictor variable and one outcome variable.
- A multiple linear regression includes two or more predictor variables and one outcome variable.
Comparison tests usually compare the means of groups. These may be the means of different groups within a sample (e.g., a treatment and control group), the means of one sample group taken at different times (e.g., pretest and posttest scores), or a sample mean and a population mean.
- A t test is for exactly 1 or 2 groups when the sample is small (30 or less).
- A z test is for exactly 1 or 2 groups when the sample is large.
- An ANOVA is for 3 or more groups.
The z and t tests have subtypes based on the number and types of samples and the hypotheses:
- If you have only one sample that you want to compare to a population mean, use a one-sample test .
- If you have paired measurements (within-subjects design), use a dependent (paired) samples test .
- If you have completely separate measurements from two unmatched groups (between-subjects design), use an independent (unpaired) samples test .
- If you expect a difference between groups in a specific direction, use a one-tailed test .
- If you don’t have any expectations for the direction of a difference between groups, use a two-tailed test .
The only parametric correlation test is Pearson’s r . The correlation coefficient ( r ) tells you the strength of a linear relationship between two quantitative variables.
However, to test whether the correlation in the sample is strong enough to be important in the population, you also need to perform a significance test of the correlation coefficient, usually a t test, to obtain a p value. This test uses your sample size to calculate how much the correlation coefficient differs from zero in the population.
You use a dependent-samples, one-tailed t test to assess whether the meditation exercise significantly improved math test scores. The test gives you:
- a t value (test statistic) of 3.00
- a p value of 0.0028
Although Pearson’s r is a test statistic, it doesn’t tell you anything about how significant the correlation is in the population. You also need to test whether this sample correlation coefficient is large enough to demonstrate a correlation in the population.
A t test can also determine how significantly a correlation coefficient differs from zero based on sample size. Since you expect a positive correlation between parental income and GPA, you use a one-sample, one-tailed t test. The t test gives you:
- a t value of 3.08
- a p value of 0.001
The final step of statistical analysis is interpreting your results.
Statistical significance
In hypothesis testing, statistical significance is the main criterion for forming conclusions. You compare your p value to a set significance level (usually 0.05) to decide whether your results are statistically significant or non-significant.
Statistically significant results are considered unlikely to have arisen solely due to chance. There is only a very low chance of such a result occurring if the null hypothesis is true in the population.
This means that you believe the meditation intervention, rather than random factors, directly caused the increase in test scores. Example: Interpret your results (correlational study) You compare your p value of 0.001 to your significance threshold of 0.05. With a p value under this threshold, you can reject the null hypothesis. This indicates a statistically significant correlation between parental income and GPA in male college students.
Note that correlation doesn’t always mean causation, because there are often many underlying factors contributing to a complex variable like GPA. Even if one variable is related to another, this may be because of a third variable influencing both of them, or indirect links between the two variables.
Effect size
A statistically significant result doesn’t necessarily mean that there are important real life applications or clinical outcomes for a finding.
In contrast, the effect size indicates the practical significance of your results. It’s important to report effect sizes along with your inferential statistics for a complete picture of your results. You should also report interval estimates of effect sizes if you’re writing an APA style paper .
With a Cohen’s d of 0.72, there’s medium to high practical significance to your finding that the meditation exercise improved test scores. Example: Effect size (correlational study) To determine the effect size of the correlation coefficient, you compare your Pearson’s r value to Cohen’s effect size criteria.
Decision errors
Type I and Type II errors are mistakes made in research conclusions. A Type I error means rejecting the null hypothesis when it’s actually true, while a Type II error means failing to reject the null hypothesis when it’s false.
You can aim to minimise the risk of these errors by selecting an optimal significance level and ensuring high power . However, there’s a trade-off between the two errors, so a fine balance is necessary.
Frequentist versus Bayesian statistics
Traditionally, frequentist statistics emphasises null hypothesis significance testing and always starts with the assumption of a true null hypothesis.
However, Bayesian statistics has grown in popularity as an alternative approach in the last few decades. In this approach, you use previous research to continually update your hypotheses based on your expectations and observations.
Bayes factor compares the relative strength of evidence for the null versus the alternative hypothesis rather than making a conclusion about rejecting the null hypothesis or not.
Hypothesis testing is a formal procedure for investigating our ideas about the world using statistics. It is used by scientists to test specific predictions, called hypotheses , by calculating how likely it is that a pattern or relationship between variables could have arisen by chance.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts, and meanings, use qualitative methods .
- If you want to analyse a large amount of readily available data, use secondary data. If you want data specific to your purposes with control over how they are generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Statistical analysis is the main method for analyzing quantitative research data . It uses probabilities and models to test predictions about a population from sample data.
Is this article helpful?
Other students also liked, a quick guide to experimental design | 5 steps & examples, controlled experiments | methods & examples of control, between-subjects design | examples, pros & cons, more interesting articles.
- Central Limit Theorem | Formula, Definition & Examples
- Central Tendency | Understanding the Mean, Median & Mode
- Correlation Coefficient | Types, Formulas & Examples
- Descriptive Statistics | Definitions, Types, Examples
- How to Calculate Standard Deviation (Guide) | Calculator & Examples
- How to Calculate Variance | Calculator, Analysis & Examples
- How to Find Degrees of Freedom | Definition & Formula
- How to Find Interquartile Range (IQR) | Calculator & Examples
- How to Find Outliers | Meaning, Formula & Examples
- How to Find the Geometric Mean | Calculator & Formula
- How to Find the Mean | Definition, Examples & Calculator
- How to Find the Median | Definition, Examples & Calculator
- How to Find the Range of a Data Set | Calculator & Formula
- Inferential Statistics | An Easy Introduction & Examples
- Levels of measurement: Nominal, ordinal, interval, ratio
- Missing Data | Types, Explanation, & Imputation
- Normal Distribution | Examples, Formulas, & Uses
- Null and Alternative Hypotheses | Definitions & Examples
- Poisson Distributions | Definition, Formula & Examples
- Skewness | Definition, Examples & Formula
- T-Distribution | What It Is and How To Use It (With Examples)
- The Standard Normal Distribution | Calculator, Examples & Uses
- Type I & Type II Errors | Differences, Examples, Visualizations
- Understanding Confidence Intervals | Easy Examples & Formulas
- Variability | Calculating Range, IQR, Variance, Standard Deviation
- What is Effect Size and Why Does It Matter? (Examples)
- What Is Interval Data? | Examples & Definition
- What Is Nominal Data? | Examples & Definition
- What Is Ordinal Data? | Examples & Definition
- What Is Ratio Data? | Examples & Definition
- What Is the Mode in Statistics? | Definition, Examples & Calculator
Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
A statistical analysis of the novel coronavirus (COVID-19) in Italy and Spain
Roles Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing
* E-mail: [email protected]
Affiliation School of Statistics, Renmin University of China, Beijing, China
- Jeffrey Chu

- Published: March 25, 2021
- https://doi.org/10.1371/journal.pone.0249037
- Reader Comments
The novel coronavirus (COVID-19) that was first reported at the end of 2019 has impacted almost every aspect of life as we know it. This paper focuses on the incidence of the disease in Italy and Spain—two of the first and most affected European countries. Using two simple mathematical epidemiological models—the Susceptible-Infectious-Recovered model and the log-linear regression model, we model the daily and cumulative incidence of COVID-19 in the two countries during the early stage of the outbreak, and compute estimates for basic measures of the infectiousness of the disease including the basic reproduction number, growth rate, and doubling time. Estimates of the basic reproduction number were found to be larger than 1 in both countries, with values being between 2 and 3 for Italy, and 2.5 and 4 for Spain. Estimates were also computed for the more dynamic effective reproduction number, which showed that since the first cases were confirmed in the respective countries the severity has generally been decreasing. The predictive ability of the log-linear regression model was found to give a better fit and simple estimates of the daily incidence for both countries were computed.
Citation: Chu J (2021) A statistical analysis of the novel coronavirus (COVID-19) in Italy and Spain. PLoS ONE 16(3): e0249037. https://doi.org/10.1371/journal.pone.0249037
Editor: Abdallah M. Samy, Faculty of Science, Ain Shams University (ASU), EGYPT
Received: July 14, 2020; Accepted: March 9, 2021; Published: March 25, 2021
Copyright: © 2021 Jeffrey Chu. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: The raw data files for the incidence of COVID-19 in Italy and Spain are available from the following links: https://github.com/pcm-dpc/COVID-19 https://github.com/datadista/datasets/tree/master/COVID%2019 .
Funding: The author(s) received no specific funding for this work.
Competing interests: The authors have declared that no competing interests exist.
Introduction
The novel coronavirus (COVID-19) was widely reported to have first been detected in Wuhan (Hebei province, China) in December 2019. After the initial outbreak, COVID-19 continued to spread to all provinces in China and very quickly spread to other countries within and outside of Asia. At present, over 45 million cases of infected individuals have been confirmed in over 180 countries with in excess of 1 million deaths [ 1 ]. Although the foundations of this disease are very similar to the severe acute respiratory syndrome (SARS) virus that took hold of Asia in 2003, it is shown to spread much more easily and there currently exists no vaccine.
Since the first confirmed cases were reported in China, much of the literature has focused on the outbreak in China including the transmission of the disease, the risk factors of infection, and the biological properties of the virus—see for example key literature such as [ 2 – 6 ]. However, more recent literature has started to cover an increasing number of regions outside of China.
For example, studies covering the wider Asia region include: investigations into the outbreak on board the Diamond Princess cruise ship in Japan, using a Bayesian framework with a Hamiltonian Monte Carlo algorithm [ 7 ]; estimation of the ascertainment rate in Japan using a Poisson process [ 8 ]; modelling the evolution of the basic and effective reproduction numbers in South Korea using Susceptible-Infected-Susceptible models [ 9 ] and generalised growth models with varying growth rates [ 10 ]; modelling the basic reproduction number in India with a classical Susceptible-Exposed-Infectious-Recovered-type compartmental model [ 11 ]; forecasting numbers of cases in Indian states using deep learning-based models [ 12 ].
Analyses on North and South America have also used similar classical methods, for example [ 13 ] model the progression of the outbreak in the United States until the end of 2021 with the simple Susceptible-Infected-Recovered model, and [ 14 ] predict epidemic trends in Brazil and Peru using a logistic growth model and machine learning techniques. However, other studies include: analysis of the spatial variability of the incidence in the United States using spatial lag and error models, and geographically weighted regression [ 15 ]; estimation of the number of deaths in the United States using a modified logistic fault-dependent detection model [ 16 ]; estimating prevalence and infection rates across different states in the United States using a sample selection model [ 17 ]; investigating the relationship between social media communication and the incidence in Colombia using non-linear regression models.
Focusing on Africa, [ 18 ] simulate and predict the spread of the disease in South Africa, Egypt, Algeria, Nigeria, Senegal, and Kenya, using a modified Susceptible-Exposed-Infectious-Recovered model; [ 19 ] apply a six-compartmental model to model the transmission in South Africa; [ 20 ] predict the spread of the disease in West Africa using a deterministic Susceptible-Exposed-Infectious-Recovered model; [ 21 ] implement Autoregressive Integrated Moving Average models to forecast the prevalence of COVID-19 in East Africa; [ 22 ] predict the spread of the disease using travel history and personal contact in Nigeria through ordinary least squares regression; [ 23 ] use logistic growth and Susceptible-Infected-Recovered models to generate real-time forecasts of daily confirmed cases in Saudi Arabia.
Aside from many of the classical models mentioned above, recent developments in the econometrics and statistics literature have led to a number of new models that could potentially be applied in the modelling of infectious diseases. These include (but are not limited to) mixed frequency analysis, model selection and combination, and dynamic time warping. Mixed frequency analysis is an iterative approach proposed for dealing with the joint dynamics of time series data which are sampled at different frequencies [ 24 ]. In the economic literature, the common example is quarterly gross domestic product (GDP) and monthly inflation. [ 25 ] notes that studying the co-movements between mixed frequency data usually involves analysing the joint process sampled at a common low frequency, however, this can mis-specify the relationship. [ 24 , 25 ] propose vector autoregressive models for mixed frequency analysis that operate at the highest sampling frequency of all the time series in the model. These models allow for the modelling of the joint dynamics of the dependent and independent variables using time disaggregation, where the low frequency variables are interpolated and time-aggregated into a higher frequency. In the context of infectious diseases, such models could be beneficial for modelling the relationship between higher frequency data such as the number of daily cases or deaths and lower frequency data relating to, say, weekly cases or deaths, news and information about health prevention measures, etc. [ 26 , 27 ] propose the use of Bayesian Predictive Synthesis (BPS) for model selection and combination. They note that there are many scenarios that generate multiple, interrelated time series, where the dependence has a significant impact on decisions, policies, and their outcomes. In addition, methods need to learn and integrate information about forecasters and models, bias, etc. and how they change over time, to improve their accuracy [ 26 ]. Decision and policy makers often use multiple sources, models, and forecasters to generate forecasts, in particular, probabilistic density forecasts. However, although complex estimation methods may have useful properties for policy makers, large standard deviations may be a result of the complexity of the data, model, etc., and it may be difficult to know the source. The aim is to use the dependencies between time series to improve forecasts over multiple horizons for policy decisions [ 27 ]. For example, in the economic literature, setting interest rates based on utility or loss that account for inflation, real economy measures, employment, etc. BPS relates to a decision maker that accounts for multiple models as providers of “forecast data” to be used for prior-posterior updating. The decision maker learns over time about relationships between agents, forecasts, and dependencies, which are incorporated into the model, and dynamically calibrate, learn, and update weights for ranges of forecasts from dynamic models, with multiple lags and predictors [ 26 ]. In epidemiology, BPS could potentially be used in a similar context to analyse the dependency between various interrelated time series such as daily cases and deaths, hospital capacity, number vaccinations, etc. Different models and sources of data could then be combined and characterised in one single model improving the accuracy of forecasts. Dynamic time warping as noted by [ 28 , 29 ] is a technique that has not been widely used outside of speech and gesture recognition. It can be used to identify the relation structure between two time series by describing their non-linear alignment with warping paths [ 28 ]. The procedure involves a local cost measure characterising the sum of the differences between pairs of realisations of data at each time point, where an optimal warping path gives the lowest total cost. The optimal path is found under a variable lead-lag structure, where the most suitable lag can then be found [ 28 ]. This then reveals and identifies the lead-lag effects between the time series data. Indeed, dynamic time warping has recently been used in the modelling of COVID-19 by [ 30 ]. [ 30 ] use the method to determine the lead-lag relation between the cumulative number of daily cases of COVID-19 in various countries, in addition to forecasting the future incidence in selected countries. This allows for the classification of countries as being in the early, middle, and late stages of an outbreak.
Controlling an infectious disease such as COVID-19 is an important, time-critical but difficult issue. The health of the global population is, perhaps, the most important factor as research is directed towards vaccines and governments scramble to implement public health measures to reduce the spread of the disease. In most countries around the world, these measures have come in the form of local or national lockdowns where individuals are advised or required to remain at home unless they have good reason not to—e.g. for educational or medical purposes, or if they are unable to work from home. However, the implications of trying to control COVID-19 are being felt not only by the health sector, but also in areas such as the economy, environment, and society.
As the number of cases of infected individuals has risen rapidly, there has been an increase in pressure on medical services as healthcare providers seek to test and diagnose infected individuals, in addition to the normal load of medical services that are offered in general. In many cases, trying to control COVID-19 has led to a backlog for and deprivation of other medical procedures [ 31 ], with healthcare providers needing to find a balance between the two. [ 32 ] note that this conflict may change the nature of healthcare with public and private health sectors working together more often. The implementation of restrictions on the movement of individuals has also led to many suggesting that anxiety and distress may lead to increased psychiatric disorders. These may be related to suicidal behaviour and morbidity and may have a long-term negative impact on the mental health of individuals [ 33 , 34 ].
In addition to restrictions on the movement of individuals, governments have required most non-essential businesses to close. This has negatively impacted national economies with many businesses permanently closing leading to a significant increase in unemployment. Limits on travel have severely affected the tourism and travel industries, and countries and economies that are dependent on these for income. Whilst many of the implications of controlling COVID-19 on the economy are negative, there have been some positive changes as businesses adapt to the ‘new normal’. For example, the banking industry is dealing with increased credit risks, while the insurance industry is developing more digital products and pandemic-focused solutions [ 32 ]. The automotive industry is expected to see profits reduced by approximately $100 billion, which may be offset by the development of software subscription services of modern vehicles [ 32 ]. Some traditional office-based businesses have been able to reduce costs by shifting to remote working, while the restaurant industry has shifted towards takeaway and delivery services [ 32 ].
In terms of the environment, the limitations on businesses that have been able to continue operating throughout the epidemic has led to possible improvements in the environment—mainly from the reduction in pollution [ 35 ]. However, societal issues have been exacerbated. [ 32 ] note that the reduction in the labour force that has resulted from controlling for COVID-19 has affected ethnic minorities and women most significantly. Furthermore, in many countries health services employ more women than men creating a dilemma for working mothers—either leave the labour force and provide childcare for their families or remain in employment and pay extra costs for childcare.
In Europe, Italy and Spain were two of the first European countries to be significantly affected by COVID-19. However, the majority of the literature covering the two countries focuses on the clinical aspects of the disease, [ 36 – 40 ], with only a limited number exploring the prevalence of the disease, [ 41 – 43 ].
As as a result of this on going pandemic, new results and reports are being produced and published daily. Thus, our motivation stems from wanting to contribute to the statistical analysis of the incidence of COVID-19 in Italy and Spain, where the literature is limited. The main contributions of this paper are: i) to model the incidence of COVID-19 in Italy and Spain using simple mathematical models in epidemiology; ii) to provide estimates of basic measures of the infectiousness and severity of COVID-19 in Italy and Spain; iii) to investigate the predictive ability of simple mathematical models and provide simple forecasts for the future incidence of COVID-19 in Italy and Spain.
The contents of this paper are organised as follows. In the data section, we describe the incidence data used in the main analysis and provide a brief summary analysis. The method section outlines the Susceptible-Infectious-Recovered model and the log-linear model used to model the incidence of COVID-19, and introduces the basic reproduction number and effective reproduction number as measures of the infectiousness of diseases. In the results section, we present the main results for the fitted models and estimates of the measures of infectiousness, in addition to simple predictions for the future incidence of COVID-19. Some concluding remarks are given in the conclusion.
The data used in this analysis consists of the daily and cumulative incidence (confirmed cases) of COVID-19 for Italy and Spain (nationally), and their respective regions or autonomous provinces. For Italy, this data covers 21 regions for 37 days from 21st February 2020 to 28th March 2020, inclusive; for Spain, this data covers 19 regions for 34 days from 27th February to 31st March 2020, inclusive. The data for Italy was obtained from [ 44 ] where the raw data was sourced from the Italian Department of Civil Protection; the data for Spain was obtained from [ 45 ] where the raw data was sourced from the Spanish Ministry of Health. The starting dates for both sets of data indicate the dates on which the first cases were confirmed in each country, however, it should be noted that in some regions cases were not confirmed until after these dates. These particular time periods were chosen as they cover over one month since the initial outbreaks in both countries and were the most up to date data available at the time of writing. In the remainder of this section, we provide a simple exploratory analysis of the incidence data.
Fig 1 plots the daily cumulative incidence for Italy and its 21 regions over the whole sample period. All cumulative incidence appears to show an exponential trend, increasing slowly for the first 14 days after the first cases are confirmed before growing rapidly. Checking the same plot on a log-linear scale, shown in Fig 2 , we find that the logarithm of cumulative incidence in some regions exhibits an approximate linear trend suggesting that cumulative incidence is growing exponentially. However, in the majority of regions (and nationally) this trend is not exactly linear, suggesting a slightly sub-exponential growth in cumulative incidence.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pone.0249037.g001
https://doi.org/10.1371/journal.pone.0249037.g002
Of all the regions in Italy, the northern region of Lombardy is one of the worst affected and Fig 3 plots the daily incremental incidence for both Lombardy and Italy, respectively. In terms of the number of new cases confirmed each day, the trends are very similar and, again, possibly exponential until peaking around 21st March 2020 before levelling off. Comparing the trends for the other regions in Fig 4 , it can be seen that other significantly affected northern regions such as Piedmont and Emilia-Romagna exhibit similarities to Lombardy—growing, peaking, and levelling around the same times. However, many other regions show some slight differences such as peaking at earlier or later dates, and even exhibiting an erratic trend.
https://doi.org/10.1371/journal.pone.0249037.g003
https://doi.org/10.1371/journal.pone.0249037.g004
In Fig 5 , things are put in perspective when the cumulative incidence of all Italian regions are plotted on the same scale. It is clear that Lombardy is the most affected region contributing to the largest share of national cumulative incidence, and indeed it is the epicentre of the outbreak in Italy.
https://doi.org/10.1371/journal.pone.0249037.g005
In the case of Spain, Fig 6 plots the daily cumulative incidence nationally and for all 19 Spanish regions over the whole sample period. The trend appears to be exponential and is similar between regions, but is also similar to that of the daily cumulative incidence in Italy. On a log-linear scale, in Fig 7 , the growth of the daily cumulative incidence appears to be closer to an exponential trend compared with Italy, due to the plots arguably exhibiting a more linear trend. It can be seen that there is a slight difference with Italy in that it appears as though most Spanish regions were affected at approximately the same time—when the country’s first cases were confirmed. This is reflected by the majority of plots starting from the very left of the x-axis, with the exception of the plots for a few regions such as Ceuta and Melilla. In Italy only a small number of regions were affected when the country’s first cases were confirmed, with the growth in cumulative incidence for the majority of the other regions coming later on.
https://doi.org/10.1371/journal.pone.0249037.g006
https://doi.org/10.1371/journal.pone.0249037.g007
The worst affected regions in Spain are Madrid and Catalonia, and Fig 8 plots the daily incremental incidence for both regions and the national trend. The growth in daily incidence, in all three cases, could be classed as being approximately exponential, however, daily incidence appears to peak on 26th March 2020 before falling and peaking again on 31st March 2020. It is confirmed that the true peak daily incidence does indeed occur on 31st March 2020 and we return to this point later on in the analysis. In comparison to other Spanish regions, it seems that Madrid and Catalonia are the exceptions as the majority of regions exhibit an exponential rise in daily incidence and peak around 26th and 27th March 2020 before falling.
https://doi.org/10.1371/journal.pone.0249037.g008
Plotting the daily incidence of all regions on the same scale in Fig 9 , it is clear that Madrid and Catalonia are the most affected regions contributing the largest share of the national cumulative incidence. Whilst Madrid and Catalonia are the main epicentres of the outbreak in Spain, many coastal regions also show significant numbers of confirmed cases, although not quite on the same scale.
https://doi.org/10.1371/journal.pone.0249037.g009
The SIR (Susceptible-Infectious-Recovered) model
In the mathematical modelling of infectious diseases, there exist many compartmental models that can be used to describe the spread of a disease within a population. One of the simplest models is the SIR (Susceptible-Infectious-Recovered) model proposed by [ 46 ], in which the population is split into three groups or compartments: those who are susceptible ( S ) but not yet infected with the disease; those who are infectious ( I ); those who have recovered ( R ) and are immune to the disease or who have deceased.
The SIR model has been extensively researched and applied in practice, thus it would not be practical to mention and cover all of the literature. However, some of the most prominent literature covers areas such as the stability and optimality of the simple SIR model ([ 47 – 51 ]); pulse vaccination strategy in the SIR model ([ 52 – 55 ]); applications of the SIR in the modelling of infectious diseases ([ 56 – 64 ]).
With regards to COVID-19, many have applied the basic SIR model (or slightly modified versions) to model the outbreak. Some particular examples include (but are not limited to): [ 2 ] who estimate the overall symptomatic case fatality risk of COVID-19 in Wuhan and use the SIR model to generate simulations of the COVID-19 outbreak in Wuhan; [ 65 ] who apply a modified SIR model to identify contagion, recovery, and death rates of COVID-19 in Italy; [ 66 ] who combine the SIR model with probabilistic and statistical methods to estimate the true number of infected individuals in France; [ 67 ] who use a number of methods including the SIR model to estimate the basic and controlled reproduction numbers for the COVID-19 outbreak in Wuhan, China; [ 68 ] who show that the basic SIR model performs better than extended versions in modelling confirmed cases of COVID-19 and present predictions for cases after the lockdown of Wuhan, China; [ 69 ] who model the temporal dynamics of COVID-19 in China, Italy, and France, and find that although the rate of recovery appears to be similar in the three countries, infection and death rates are more variable; [ 70 ] who simulate the outbreak in Wuhan, China, using an extended SIR model and investigate the age distribution of cases; [ 71 ] who study the number of infections and deaths from COVID-19 in Sweden using the SIR model; [ 72 ] who use the SIR model, with an additional parameter for social distancing, to model and forecast the early stages of the COVID-19 outbreak in Brazil.
In reference to the SIR model, [ 74 ] note that it “examines only the temporal dynamics of the infection cycle and should thus be appropriate for the description of a well-localised epidemic outburst”, therefore, it would appear to be reasonable for use in analysis at city, province, or country level. In the form above, the dynamics of the model are controlled by the parameters β and γ , representing the rates of transition from S to I (susceptibility to infection), and I to R (infection to recovery or death), respectively.
To fit the model and find the optimal parameter values of β and γ , we use the optim function in R [ 75 ] to solve the minimisation problem. The system of differential equations, Eqs ( 1 ) to ( 3 ), are set up as a single function. The model is then initialised with starting values for S , I , and R , with parameters β and γ unknown. We obtain the daily cumulative incidence for the sample period, total population ( N ), and the susceptible population ( S ) as the total population minus the number of currently infected individuals. This is defined as the cumulative number of infected individuals minus the number of recovered or dead, however, these exact values are difficult to obtain. Thus, the cumulative number of infected individuals at the start date of the sample period is used as a proxy—since at the start date of the disease, this is likely to be close to the true value, as the number of recovered or dead should be very small (if not zero).
The residual sum of squares is then defined and set up as a function of β and γ . The optim package is used for general purpose optimisation problems, and in this case it is used to minimise the function RSS with respect to the sample of cumulative incidence. More specifically, we use the limited memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS-B) algorithm for the minimisation, which allows us to specify box constraints (lower and upper bounds) for the unknown parameters β and γ . The lower and upper bounds of zero and one, respectively, were selected for both parameters. The optim function then searches for the β and γ that minimise the RSS function, given starting values of 0.5 for both parameters. The optimal solution is found via the gradient method by repeatedly improving the estimates of RSS to try and find a solution with a lower value. The function makes small changes to the parameters in the direction of where RSS changes the fastest, where in this direction the lowest value of RSS is. This is repeated until no further improvement can be made or the improvement is below a threshold.
We consider convergence as the main criteria for finding an optimal solution in the minimisation of RSS —when the lowest RSS has been found, and no further improvement can be found or the improvement is below a threshold. In the case where convergence is not achieved, or there is some related error, then we use the parscale function in the optimisation. As the true values of β and γ are unknown, in the default case, the parameters are adjusted by a fixed step starting from their initial values. Most common issues were addressed using the parscale function to rescale—alter the sensitivity/magnitude of the parameters on the objective function. In other words, it allows the algorithm to compute the gradient at a finer scale (similar to the ndeps parameter—used to adjust step sizes for the finite-difference approximation to the gradient). In most cases, issues were solved by using a step size of 10 −4 . Of course, smaller step sizes could be used, but there is a risk that selecting too small a step size will lead to the optimal values of β and γ being found at their starting values. However, the results should be interpreted with caution. It is possible that estimates will vary with different population sizes N and the starting values specified for β and γ , which may also cause the optimisation process to be unstable.
It should be noted that the application of the basic SIR model to COVID-19 simplifies the analysis and makes the strong assumption that individuals who become infected but recover are immune to COVID-19. This is assumed purely for the simplification of modelling and we do not claim this to be true in reality. At present, it remains unclear whether those who recover from infection are immune [ 76 ]. Indeed, there have been studies and unconfirmed reports of individuals who have possibly recovered but then subsequently tested positive for the virus again, see for example [ 77 – 79 ].
The basic reproduction number R 0
Whilst the fitted model and optimal parameters allow us to make a simple prediction about how the trajectory of the number of susceptible, infectious, and recovered individuals evolves over time, a more useful statistic or parameter that can be computed from the fitted model is the basic reproduction number R 0 . Originally developed for the study of demographics in the early 20th century, it was adapted for use in the study of infectious diseases in the 1950’s [ 80 ]. It is defined as the “expected number of secondary infections arising from a single individual during his or her entire infectious period, in a population of susceptibles” [ 80 ], and is widely considered to be a fundamental concept in the study of epidemiology. In other words, it is the estimated number of people that an individual will go on to infect after becoming infected.
The R 0 value can provide an indication of the severity of the outbreak of an infectious disease: if R 0 < 1, each infected individual will go on to infect less than one individual (on average) and the disease will die out; if R 0 = 1, each infected individual will go on to infect one individual (on average) and the disease will continue to spread but will be stable; if R 0 > 1, each infected individual will go on to infect more than one individual (on average) and the disease will continue to spread and grow, with the possibility of becoming a pandemic ([ 80 , 81 ]).
Log-linear model
https://doi.org/10.1371/journal.pone.0249037.g010
To fit the log-linear model, we use the incidence package [ 82 ] in R [ 75 ] to obtain the optimal values of the parameters. Using the estimated parameters, the fitted model can be used to predict the trajectory of the incidence up until the peak incidence in the growth phase. However, although the log-linear model allows for the modelling and prediction of the incidence, compared with the SIR model it does not provide any indication about the number of susceptible or recovered individuals.
We are able to use the epitrix R package [ 84 ] to implement the method by [ 83 ] for empirical distributions to estimate R 0 from the growth rate r . However, [ 83 ] note that an “epidemic model implicitly specifies a generation interval distribution” (also known as the serial interval distribution), which is defined as “the time between the onset of symptoms in a primary case and the onset of symptoms in secondary cases” [ 85 ]. As we do not have access to more detailed COVID-19 patient data, we are not able to compute the parameters of the serial interval distribution directly. However, a number of existing analyses of COVID-19 patient data report some preliminary estimates of the best fitting serial interval distributions and their corresponding model parameters. These are: i) gamma distribution with mean μ = 7.5 and standard deviation σ = 3.4 [ 81 ]; ii) gamma distribution with mean μ = 7 and standard deviation σ = 4.5 [ 2 ]; iii) gamma distribution with mean μ = 6.3 and standard deviation σ = 4.2 [ 86 ]. By using these three serial intervals in conjunction with the above method, we are able to obtain estimates of R 0 from estimates of the growth rate r . It should be noted that serial interval distributions are not only restricted to the gamma distribution—other common distributions used include the Weibull and log-normal distributions, and that the parameters are dependent on a number of factors including the time to isolation [ 86 ].
The effective reproduction number R e
As mentioned above, the estimation of the R 0 value is not always ideal, due to it being a single fixed value reflecting a specific period of growth (in the log-linear model) or requiring assumptions that only hold true in specific time periods (in the basic SIR model). In other words, it is “time and situation specific” [ 85 ]. In reality, the reproduction number will vary over time but it will also be influenced by governments and health authorities implementing measures in order to reduce the impact of the disease. Therefore, a more useful approach for measuring the severity of an infectious disease is to track the reproduction number over time. The effective reproduction number R e is one way to achieve this, and thus allows us to see how the reproduction number changes over time in response to the development of the disease itself but also effectiveness of interventions. Although there are numerous methods that can be used to analyse the severity of a disease over time, the majority are not straightforward to implement (especially in software) [ 85 ].
One popular method for estimating R e is that proposed by [ 85 ]. The basic premise of this method is that “once infected, individuals have an infectivity profile given by a probability distribution w s , dependent on time since infection of the case, s , but independent of calendar time, t . For example, an individual will be most infectious at time s when w s is the largest. The distribution w s typically depends on individual biological factors such as pathogen shedding or symptom severity” [ 85 ].
The function models the transmissibility of a disease with a Poisson process, such that an individual infected at time t − s will generate new infections at time t at a rate of R t w s , where R t is the instantaneous (effective) reproduction number at time t . Thus, the incidence at time t is defined to be Poisson distributed with mean equal to the average daily incidence (number of new cases) at time t . This value is just for a single time period t , however, estimates for a single time period can be highly variable meaning that it is not easy to interpret, especially for making policy decisions. Therefore, we consider longer time periods of one week (seven days)—assuming that within a rolling window the instantaneous reproduction number remains constant. Note that there is a potential trade off, as using longer rolling windows gives more precise estimates of R t but this means fewer estimates can be computed (requires more incidence values to start with) and a more delayed trend reducing the ability to detect changes in transmissibility. Whereas shorter rolling windows lead to more rapid detection in changes but with more noise. Using this method, it is recommended that a minimum cumulative daily incidence of 12 cases have been observed before attempting to estimate R e . For the data sets used, this does not pose a problem as a cumulative total of 16 and 17 cases, respectively, exist on the first day of the sample at the country level, and by the seventh day the totals are around 200 and 650 for Spain and Italy, respectively.
From the posterior distribution, the posterior mean R t , τ can be computed at time t for the rolling window of [ t − τ , t ] by the ratio of the gamma distribution parameters. We refer the readers to the supplementary information of [ 85 ] for further details regarding the Bayesian framework. As noted by [ 85 ], this method works best when times of infection are known and the infectivity profile or distribution can be estimated from patient level data. However, as mentioned above, we do not have access to this level of data, and instead utilise three different serial intervals from the literature that have been estimated from real data.
In practice, the transmission of a disease will vary over time especially when health prevention measures are implemented. However, this method is the only reproduction number that can be easily computed in real-time, and in comparison to similar methods, it captures the effect of control measures since it will cause sudden decreases in estimates compared with other methods.
In this analysis, we use the most basic version of this method and estimate the effective reproduction number over a rolling window of seven days. This appears to be sufficient and in line with our results, as we do not suffer from the problem of small sample sizes as the samples are sufficiently large and we start computing the effective reproduction number after one mean serial interval. It should be noted that estimates of this reproduction number are dependent on the distribution of the infectiousness profile w s . In addition, it is known that this distribution may not always be well documented, especially in the early parts of an epidemic. However, here we assume that the serial interval is defined for our sample period and the use of the three serial intervals from the literature appears to give satisfactory results.
If problems did arise, or to account for uncertainty in the serial interval distribution, an alternative method is to implement a modified procedure by [ 85 ], which allows for uncertainty in the serial interval distribution. This modified method assumes that the serial interval is gamma distributed but the mean and standard deviation are allowed to vary according to a standard normal distribution. Some N * pairs of means and standard deviations are simulated—mean first and standard deviation second, with the constraint that the mean is less than the standard deviation to ensure that for each pair the probability density function of the serial interval distribution is null at time t = 0. Then, for each rolling window 1000 realisations are sampled of the instantaneous reproduction number using the posterior distribution conditional on the pair of parameters.
The SIR model and R 0
For both Italy and Spain, we set up and solve the minimisation problem for the SIR model described in Section for region-level and national-level COVID-19 incidence for the first 14 days after the first cases were confirmed in each respective country and region. The first 14 days after the first cases are detected can be considered to be the early stage of an outbreak, and it is reasonable to assume that there are few, if no, infected or immune individuals prior to this. However, it is a rather strong assumption as it is possible that individuals may be infected but do not display any symptoms. Tables 1 and 2 show the output corresponding to each region/country including the date that the first cases were confirmed, the population size (obtained from [ 88 ]), the cumulative number of cases at the 14th day after the first cases were confirmed, the fitted estimates for the parameters β and γ , and estimates for R 0 .
https://doi.org/10.1371/journal.pone.0249037.t001
https://doi.org/10.1371/journal.pone.0249037.t002
From Tables 1 and 2 , we observe that many of the first regions to be affected in both countries are those with the largest population sizes, however, the cumulative number of cases (after the first 14 days) in these regions are not always the highest among all regions. The estimates of the parameters β and γ also do not show any particular trends and this is reflected in the estimated R 0 values. It can be seen that for all regions in both Italy and Spain, the estimated R 0 values fall between one and three. This suggests that, according to the thresholds described above, the disease is spreading and growing in all Italian and Spanish regions during the 14 days after the first localised cases were confirmed. At a national level, the estimated values of R 0 are greater than two for both countries, again, suggesting a spreading and growing disease. This is perhaps not surprising since this time period reflects the early stages of the spread of the disease, thus we would expect it to be growing and spreading quickly before any preventative action is taken.
We note that in Tables 1 and 2 , there are some cases where the estimated value of β is very close to or at the upper limit of 1.000—e.g. Lombardy (Italy) and Madrid (Spain). This leads to the consequence that the parameter estimates appear to be bound by the upper limit. However, all parameter estimates are dependent on the starting values defined for β and γ , and the upper and lower bounds specified. For all cases of estimating the parameters in Tables 1 and 2 , we used the same optimisation procedure and criteria for determining a satisfactory estimate that is the convergence in the minimisation of the RSS ( Eq (4) ). In all cases, convergence was achieved but this is still slightly problematic. For cases where the estimated value of β is 1.000, although convergence was achieved, this indicates only that it generates the lowest RSS within the upper and lower limits defined. Therefore, there may or may not exist values of the parameter outside of this range that may be more optimal. Indeed, the results may vary depending on the upper and lower bounds, and the starting values that are selected. Thus, there is also the question of how to change the starting values and bounds appropriately (instead of, say, simply increasing them). Furthermore, as the R 0 value in the SIR model is computed as β / γ , another consequence of the estimated value of β being 1.000 is that the true value of β may actually be larger than this, and so the true value of R 0 may be larger than the estimated value.
Using the estimated parameters for the best fitted models, the predicted trajectories of the numbers in each of the compartments of the model can be generated. For brevity, in the remainder of the analysis, we show only the results for Italy, Spain, and their worst affected regions. Fig 11 plots the observed and predicted cumulative incidence for the 14 days immediately following the first confirmed cases in Lombardy and Italy, respectively. It can be seen that the model appears to under predict the true total number of cases in both cases during the early part of the outbreak before over estimating towards the end of the 14 days. In Fig 12 the SIR model trajectories are plotted along with the observed cumulative incidence on a logarithmic scale for Lombardy and Italy. The under prediction of the cumulative incidence in the first 14 days (to the left of the vertical dashed black line) is indicated by the solid red line (predicted cumulative incidence) lying below the black points (observed cumulative incidence) however, after the initial 14 days and after the implementation of a nationwide lock down (vertical dashed red line), the observed cumulative incidence grows at a slower rate than predicted by the fitted model. Indeed, this reflects the fact that the model is based only on the initial 14 days and does not account for any interventions.
https://doi.org/10.1371/journal.pone.0249037.g011
https://doi.org/10.1371/journal.pone.0249037.g012
In Fig 13 , the observed and predicted cumulative incidence for the 14 days immediately following the first confirmed cases in Catalonia, Madrid, and Italy, respectively, are shown. In contrast to the results for Italy, the fitted model for all three appears to predict the true total number of cases across the whole of the first 14 days reasonably well. Fig 14 plots the SIR model trajectories and the observed cumulative incidence on a logarithmic scale for Catalonia, Madrid, and Spain. Here, the more accurate predictions of the cumulative incidence are reflected in the area to the left of the vertical dashed black line. However, it can be seen that at the time when the nationwide lock down came into force (vertical dashed red line) the growth of the true total number of cases slowed down. It is likely that this is coincidental, since it is known that the effect on the incidence of infectious diseases from health interventions is not immediate, but instead lags behind.
https://doi.org/10.1371/journal.pone.0249037.g013
https://doi.org/10.1371/journal.pone.0249037.g014
Log-linear model and R 0
Following the SIR model, we implemented the log-linear model as described above for region-level and national-level COVID-19 daily incidence for the entire growth phase (from the time of the first confirmed cases until the time at which daily incidence peaks). The estimated parameters of the fitted log-linear models for the daily incidence of Lombardy and Italy, respectively, are shown in Table 3 . It can be seen that the peak daily incidence in both Lombardy and at country level occurred on the same day (21st March 2020), however, the growth rate (doubling time) is found to be slightly greater (shorter) at country level (0.18 and 3.88) compared with the Lombardy region (0.16 and 4.34). In comparison to the SIR model and modelling the cumulative incidence, the log-linear model modelling the daily incidence in the growth phase (as shown in Fig 15 ) appears to be slightly more accurate.
Upper and lower limits of the 95% confidence intervals are indicated by the dashed red lines.
https://doi.org/10.1371/journal.pone.0249037.g015
https://doi.org/10.1371/journal.pone.0249037.t003
In Table 4 , the estimated parameters of the fitted log-linear models for the daily incidence of Madrid, Catalonia, and Spain, respectively, are given. Similarly, the peak daily incidence occurs on the same day (31st March 2020) for Madrid, Catalonia, and Spain, although this is later than that for Italy. Interestingly, the growth rate (doubling time) is greatest (shortest) for Catalonia (0.24 and 3.85), whilst Madrid and Spain share similar growth rates and doubling times (0.21/0.22 and 3.24/3.21). It should be noted that there appears to be a slight difference in the observed daily incidence compared with the case of Italy and its regions. In Fig 16 , it can be seen that the observed daily incidence appears to initially peak in the last few days of March in all cases before falling, but then increases to a higher peak at the end of the growth phase. This seems to throw off the fitted log-linear model, as after the initial (approximate) 14 days the fitted model under predicts and then over predicts the daily incidence.
https://doi.org/10.1371/journal.pone.0249037.g016
https://doi.org/10.1371/journal.pone.0249037.t004
As with the SIR model, we are also able to use the fitted log-linear models in conjunction with the three serial intervals mentioned above to compute estimates of the R 0 value. Table 5 shows the mean estimates of the R 0 value for Italy, Spain, and their most affected regions, computed from the fitted log-linear models and the three serial intervals. In each case, the mean estimates are computed from 10,000 samples of R 0 values generated from the log-linear regression of the incidence data in the growth phase, and the distributions of these samples are plotted in S1 Fig . Compared with the estimates from the SIR model, we find that in all but the case of Italy, the estimates of R 0 from the log-linear model are greater than that from the SIR model—in these cases, the lowest estimates of R 0 from the log-linear models are larger by between 0.5 to 1. In the case of Italy, we find that the estimate of R 0 computed from the SIR model is approximately the same as that computed from the log-linear model using a serial interval using a gamma distribution with mean μ = 7 and standard deviation σ = 4.5 [ 2 ]. Using the log-linear models, the largest R 0 values computed are for Catalonia, whereas the smallest values are for Lombardy. It can also be seen that serial distributions with a lower mean appear to correspond with lower R 0 values. A possible explanation for the difference between the estimated R 0 values computed from the SIR models and the log-linear models is that the only incidence data from the first 14 days was used in the former, whereas incidence data from the whole growth phase was used in the latter—almost double the data. Therefore, it is arguable that the R 0 estimates from the log-linear models could be considered to be more accurate.
https://doi.org/10.1371/journal.pone.0249037.t005
Effective reproductive number R e .
Turning towards the more dynamic measure of the infectiousness of diseases, Figs 17 and 18 plot the estimated reproductive numbers computed for Lombardy, Italy, Madrid, Catalonia, and Spain, over the entire sample period. Using the method proposed by [ 85 ], in each case estimates were computed using rolling windows of the daily incidence over the previous 7 days and the same three serial distributions as for the log-linear models. As a result, no estimates are computed for the first 7 days of each respective sample period. In all cases, we analyse and compute the R e values over the whole sample period available allowing us to see how the infectiousness of COVID-19 varies during the initial outbreak stages and the effect of any interventions implemented by the respective governments. In Fig 17 , we observe that for both Lombardy and Italy, R e is generally decreasing over the time (under any of the three serial distributions), and although it is initially larger for Italy, after approximately the first 7 days the R e values are similar. However, the trend of R e both to the left and right (before and after) of the nationwide lockdown (indicated by the dotted line) shows some differences. Prior to the nationwide lockdown, R e decreases rapidly towards a value of between three and four, which could be attributed to the fact that northern Italy (including Lombardy) was the most affected area in the early stages of the outbreak and lockdowns local to the area were already being enforced from 21st February 2020. Thus, this is likely to have contributed (in part) to the initial reduction in the R e value. After the nationwide lockdown came into force on 9th March 2020, R e continues to decrease but at a slower pace and appears to level off approximately 14 days later—this coincides with the peak in daily incidence on 21st March 2020. After this point, it is likely that the effects of the nationwide lockdown are starting to appear with R e appearing to decrease again more rapidly towards the critical value of one (solid horizontal line)—suggesting that the disease is still spreading but stabilising.
Upper and lower limits of the 95% confidence intervals for the mean are indicated by the red dashed lines, and the grey dotted line indicates the date at which the national lock down becomes effective.
https://doi.org/10.1371/journal.pone.0249037.g017
https://doi.org/10.1371/journal.pone.0249037.g018
In Fig 18 , we observe a different trend in the R e value for Madrid, Catalonia, and Spain, compared with Lombardy and Italy. Whilst R e exhibits a decrease over the sample time period (under any of the three serial distributions), the initial values are actually larger for Madrid and Catalonia, however, the values for all three are similar after the initial 7 days. The trend in the estimated R e values before and after the nationwide lockdown again show some differences, but also differ to those for the cases of Lombardy and Italy. Prior to the nationwide lockdown (indicated by the dotted line), the trend of the estimated R e values is very erratic: decreasing, increasing, and then decreasing again. This could be due to the daily incidence for Madrid, Catalonia, and Spain, showing greater variation compared with that for Italy before the respective lockdowns. It is found that in the period before the lockdowns, Spanish daily incidence appears to show more alternation between increases and decreases compared with the previous day’s incidence, whilst Italian daily incidence shows much less. After the nationwide lockdown on 14th March 2020, for all three cases the estimated R e decreases significantly towards a value of two. More specifically, in mid-March 2020 daily incidence for Madrid, Catalonia, and Spain, levels off corresponding to the reduction in R e , but in the run up to 23rd March 2020 daily incidence again becomes more variable and alternates between significantly larger and smaller daily incidence, with R e levelling off. After 23rd March 2020, this levelling off is more sustained for Madrid and Spain compared with Catalonia. This may be attributed to the daily incidence initially peaking and then decreasing much more significantly for Catalonia, leading to a more significant decrease in R e at the latter end of the sample period. In general, the estimated R e values are larger for Spain than Italy, since Spain is lagging behind in terms of the start of the outbreak, however, it is found that the estimated R e is larger for Italy than Spain, but larger for Madrid and Catalonia than Lombardy.
Predictive ability of models.
Whilst the results regarding the estimated reproduction values ( R 0 and R e ) provide useful indicators about the infectiousness of COVID-19 and the variability over time, the predictive ability of models is also key—especially in the decay phase of an outbreak after the daily incidence has peaked and is in decline. Predictions about the daily incidence in the decay phase can contribute to determining whether health interventions are working, but can additionally provide time frames for when daily incidence may reach certain thresholds—e.g. below which the disease may be considered under control. To compare the predictive ability of the SIR and log-linear models, we use the projections package [ 89 ] in R [ 75 ]. As this section acts to provide only a brief analysis of the predictive ability of the models, we refer the readers to [ 89 ] for in-depth documentation regarding the finer details of the computations. The initial step is to consider which of the two models provides the best predictive ability in the growth phase of the COVID-19 outbreak and for simplicity, we analyse only Italy and Spain at country level. Using the estimated R 0 values for Italy and Spain from the SIR and log-linear models above, we combine these with the three serial distributions mentioned earlier. We then use the projections package [ 89 ] to forecast and predict the daily incidence for Italy and Spain from the 14th day (since the first cases in each location) until the day of peak incidence.
Plots of the true daily incidence in Italy and Spain during their respective growth phases and the predicted values using the SIR and log-linear models are shown in Figs 19 and 20 . In each figure, the first row plots the predictions using the SIR model; the second row plots the predictions using the log-linear model. For the case of Italy, the plots in Fig 19 appear to show that the predictions using the R 0 value estimated from the SIR model and the serial interval of a gamma distribution with mean μ = 7.5 and standard deviation σ = 3.4 [ 81 ] provide the most accurate general predictions. However, although using the R 0 value estimated from the log-linear model generates predictions which are accurate up until the last 7 days of the growth phase (where all three cases show over prediction), these results are more consistent compared with those using the SIR model. For the case of Spain, the plots in Fig 20 show that the predictions using the R 0 value estimated from the SIR model are consistent but significantly under predicting the observed daily incidence. In contrast, predictions using the R 0 value estimated from the log-linear model are consistent and accurate up until the initial peak in daily incidence a few days before the true peak at the end of the growth phase. Based on these results for the growth phase of the outbreak, we propose to use the log-linear model to compute basic predictions for the decay phase.
95% confidence intervals for the predicted incidence are indicated by the shaded light purple regions.
https://doi.org/10.1371/journal.pone.0249037.g019
https://doi.org/10.1371/journal.pone.0249037.g020
At the time of conducting this part of the analysis, approximately one month of daily incidence data was available for the decay phase (following peak daily incidence) of both Italy and Spain. Similarly, we follow the methodology for fitting the log-linear model but now apply it to the decay phase daily incidence. The model is fitted to the decay phase daily incidence in the same way, and model parameters can be computed. Note that for the decay phase, the values and interpretation of the estimated parameters change—the growth rate takes a negative value and the doubling time becomes the halving time (both reflecting the decay and decrease in daily incidence). The fitted log-linear regressions for Italy and Spain are shown in the left hand plots of Figs 21 and 22 , respectively. The fitted models appear to provide reasonable fits to the observed decay phase daily incidence much like the case for the growth phase.
Plots of the observed (dot-dashed black line) and projected daily incidence for the next 180 days using the log-linear model and serial interval distributions SI 1 (green line), SI 2 (blue line), and SI 3 (red line) (right).
https://doi.org/10.1371/journal.pone.0249037.g021
https://doi.org/10.1371/journal.pone.0249037.g022
Also, as in the growth phase, the R 0 value can still be computed for the log-linear model during the decay phase, and for consistency we obtain mean estimates of R 0 from 10,000 samples of R 0 generated from the log-linear regressions of the daily incidence during the decay phase in conjunction with the three serial distributions. Distributions of these estimates are plotted in S2 Fig and it can be seen that (in contrast to the growth phase) the mean estimates of R 0 for Italy and Spain, individually, are very similar (under the three serial distributions)—between 0.85 and 0.87 for Italy, and 0.77 and 0.83 for Spain. Using the mean estimated R 0 values and the three serial distributions, we computed projections of the daily incidence for the 180 days immediately following the end of the decay phase sample period on 22nd April 2020. The paths of these projections for Italy and Spain are shown in the right hand plots of Figs 21 and 22 , respectively.
A simple comparison of the projected daily incidence for both countries is given in Table 6 , at one and two months following the end of the decay phase sample period. Observed daily incidence for the remainder of the decay phase was obtained from [ 44 , 90 , 91 ]. In general, it appears that the predictions for future daily incidence (under all three serial distributions) in both Italy and Spain are significantly greater than the observed daily incidence. At the one month time point (21st May 2020) projections of daily incidence for Italy are approximately twice as large as the true incidence; projections of daily incidence for Spain are approximately two to three times as large as the true incidence. Moving forward to the two month time point (21st June 2020) projections of the daily incidence for Italy are approximately two to three times as large as the true incidence; projections of the daily incidence for Spain are up to twice as large as the true incidence. However, the projection of Spanish daily incidence using the serial interval of a gamma distribution with mean μ = 6.3 and standard deviation σ = 4.2 [ 86 ] is almost identical to the true incidence.
https://doi.org/10.1371/journal.pone.0249037.t006
Whilst the results of the projections generally show significant over estimation of future daily incidence in both Italy and Spain, they do provide some additional information to the reproduction values regarding the trends of daily incidence. However, such forecasts should be not be taken directly at face value as there are a number of pitfalls that will influence the predictions. Limited decay phase incidence data was available at the time of the original analysis, which is likely to have led to less accurate estimates of R 0 and thus predictions. On a related note, the predictions are conditional on the data up until the end of the sample decay phase data and thus do not account for any health policies or interventions implemented after this, likely leading to the over estimation.
In this paper, we have provided a simple statistical analysis of the novel Coronavirus (COVID-19) outbreak in Italy and Spain—two of the worst affected countries in Europe. Using data of the daily and cumulative incidence in both countries over approximately the first month after the first cases were confirmed in each respective country, we have analysed the trends and modelled the incidence and estimated the basic reproduction value using two common approaches in epidemiology—the SIR model and a log-linear model.
Results from the SIR model showed an adequate fit to the cumulative incidence of Spain and its most affected regions in the early stages of the outbreak, however, it showed significant under estimation in the case of Italy and its most affected regions. Estimates of the basic reproduction number in the early stage of the outbreak from the model were found to be greater than one in all cases, suggesting a growing infectiousness of COVID-19—in line with expectations. Applying the log-linear regression model to the daily incidence, results for the growth phase of the outbreak in Italy and Spain revealed a greater growth rate for Spain compared with Italy (and their most affected regions)—approximately between 0.21 to 0.24 for the former and 0.15 to 0.18 for the latter. The time for the daily incidence to double for Spain was also found to be shorter than Italy (approximately three days compared to four days).
With the lack of detailed clinical COVID-19 data for the two countries, we utilised existing results regarding the serial interval distribution of COVID-19 from the literature to estimate the basic reproduction number via the log-linear model. Estimates of this value were found to be between 2.1 and 3 for Italy and its most affected region Lombardy, and between 2.5 and approximately 4 for Spain and its most affected regions of Madrid and Catalonia. Further analysis of the effective reproduction number (based on the incidence over the previous seven days) indicated that in both countries the infectious of COVID-19 was decreasing and reflecting the positive impact of health interventions such as nationwide lock downs.
Basic predictions of future daily incidence in Italy and Spain were estimated using the log-linear regression model for the decay phase of the outbreak. Estimates of the projected daily incidence at various time points in the future were generally found to be between two to three times larger than the true levels of daily incidence. These results highlight the fact that the estimates may only give reasonable indications in the short term, since they are based on past data which may or may not account for factors which change in the short term—e.g. new health interventions, public policy, etc.
Despite the simplicity of our results, we believe that they provide an interesting insight into the statistics of the COVID-19 outbreak in two of the worst affected countries in Europe. Our results appear to indicate that the log-linear model may be more suitable in modelling the incidence of COVID-19 and other infectious diseases in both the growth and decay phases, and for short term predictions of the growth (or decay) of the number of new cases when no intervention measures have recently been implemented. In addition, the results could be useful in contributing to health policy decisions or government interventions—especially in the case of a significant second wave of COVID-19. However, these results should be used in conjunction with the results from other more complex mathematical and epidemiological models.
Supporting information
S1 fig. plots of the distributions of samples of r 0 values computed from the fitted log-linear regressions of growth phase incidence..
i) Lombardy (top left); ii) Italy (top right); iii) Madrid (middle left); iv) Catalonia (middle right); v) Spain (bottom). a) SI 1 (blue); b) SI 2 (red) c) SI 3 (green).
https://doi.org/10.1371/journal.pone.0249037.s001
S2 Fig. Plots of the distributions of samples of R 0 values computed from the fitted log-linear regressions of decay phase incidence.
i) Italy (left); ii) Spain (right). a) SI 1 (green); b) SI 2 (red) c) SI 3 (blue).
https://doi.org/10.1371/journal.pone.0249037.s002
- 1. Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), 2020. Coronavirus COVID-19 (2019-nCoV). Available at: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6 .
- View Article
- Google Scholar
- PubMed/NCBI
- 13. Atkeson, A., 2020. What Will Be the Economic Impact of COVID-19 in the US? Rough Estimates of Disease Scenarios. National Bureau of Economic Research, Working Paper 26867.
- 17. Benatia, D., Godefroy, R. and Lewis, J., 2020. Estimating COVID-19 Prevalence in the United States: A Sample Selection Model Approach. Available at: https://ssrn.com/abstract=3578760 .
- 32. McKinsey & Company, 2020. COVID-19: Implications for business. Available at: https://www.mckinsey.com/business-functions/risk/our-insights/covid-19-implications-for-business .
- 44. GitHub, 2020a. pcm-dpc/COVID-19: COVID-19 Italia—Monitoraggio situazione. Available at: https://github.com/pcm-dpc/COVID-19 .
- 45. GitHub, 2020b. datasets/COVID 19 at master ⋅ datadista/datasets. Available at: https://github.com/datadista/datasets/tree/master/COVID%2019 .
- 64. Correia A.M., Mena F.C., Soares A.J., 2011. An Application of the SIR Model to the Evolution of Epidemics in Portugal. In: M. Peixoto, A. Pinto and D. Rand eds. Dynamics, Games and Science II. Springer Proceedings in Mathematics, vol 2. Berlin: Springer. pp. 247-250.
- 65. Calafiore, G.C., Novara, C. and Possieri, C., 2020. A Modified SIR Model for the COVID-19 Contagion in Italy. arXiv:2003.14391v1.
- 66. Roques, L., Klein, E., Papax, J., Sar, A. and Soubeyrand, S., 2020. Using early data to estimate the actual infection fatality ratio from COVID-19 in France (Running title: Infection fatality ratio from COVID-19). arXiv:2003.10720v3.
- 67. You, C., Deng, Y., Hu, Y., Sun, J., Lin, Q., Zhou, F., et al. Estimation of the Time-Varying Reproduction Number of COVID-19 Outbreak in China. Available at SSRN: https://ssrn.com/abstract=3539694 .
- 71. Qi, C., Karlsson, D., Sallmen, K. and Wyss, R., 2020. Model studies on the COVID-19 pandemic in Sweden. arXiv:2004.01575v1.
- 72. Bastos, S.B. and Cajuero, D.O., 2020. Modeling and forecasting the early evolution of the Covid-19 pandemic in Brazil. arXiv:2003.14288v2.
- 75. R Development Core Team, 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2020).
- 76. World Health Organization, 2020. “‘Immunity passports” in the context of COVID-19’. Available at: https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19 .
- 79. Reuters, 2020. “Explainer: Coronavirus reappears in discharged patients, raising questions in containment fight”. Available at: https://uk.reuters.com/article/us-china-health-reinfection-explainer/explainer-coronavirus-reappears-in-discharged-patients-raising-questions-in-containment-fight-idUKKCN20M124 .
- 82. Jombart, T., Kamvar, Z.N., FitzJohn, R., Cai, J., Bhatia, S., Schumacher, J, et al. 2020. incidence: Compute, Handle, Plot and Model Incidence of Dated Events. R package version 1.7.1. https://CRAN.R-project.org/package=incidence .
- 84. Jombart, T., Cori, A., Kamvar, Z.N. and Schumacher, D., 2019. epitrix: Small Helpers and Tricks for Epidemics Analysis. R package version 0.2.2. https://CRAN.R-project.org/package=epitrix .
- 87. Cori, A., Cauchemez, S., Ferguson, N.M., Fraser, C., Dahlqwist, E., Demarsh, P.A., et al. 2019. EpiEstim: Estimate Time Varying Reproduction Numbers from Epidemic Curves. R package version 2.2-1 https://cran.r-project.org/package=EpiEstim .
- 88. Eurostat, 2019. Population: demography, population projections, census, asylum & migration—Overview. Available at: https://ec.europa.eu/eurostat/web/population/overview .
- 89. Jombart, T., Nouvellat, P., Bhatia, S. and Kamvar, Z.N., 2018. projections: Project Future Case Incidence. R package version 0.3.1. https://CRAN.R-project.org/package=projections .
- 90. Worldometer, 2020. Worldometer—real time world statistics. Available at: https://www.worldometers.info/ .
- 91. Ministerio de Sanidad, Consumo y Bienestar Social. Enfermedad por nuevo coronavirus, COVID-19. Available at: https://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov-China/ .

- Search by keyword
- Search by citation
Page 1 of 3
A generalization to the log-inverse Weibull distribution and its applications in cancer research
In this paper we consider a generalization of a log-transformed version of the inverse Weibull distribution. Several theoretical properties of the distribution are studied in detail including expressions for i...
- View Full Text
Approximations of conditional probability density functions in Lebesgue spaces via mixture of experts models
Mixture of experts (MoE) models are widely applied for conditional probability density estimation problems. We demonstrate the richness of the class of MoE models by proving denseness results in Lebesgue space...
Structural properties of generalised Planck distributions
A family of generalised Planck (GP) laws is defined and its structural properties explored. Sometimes subject to parameter restrictions, a GP law is a randomly scaled gamma law; it arises as the equilibrium la...
New class of Lindley distributions: properties and applications
A new generalized class of Lindley distribution is introduced in this paper. This new class is called the T -Lindley{ Y } class of distributions, and it is generated by using the quantile functions of uniform, expon...
Tolerance intervals in statistical software and robustness under model misspecification
A tolerance interval is a statistical interval that covers at least 100 ρ % of the population of interest with a 100(1− α ) % confidence, where ρ and α are pre-specified values in (0, 1). In many scientific fields, su...
Combining assumptions and graphical network into gene expression data analysis
Analyzing gene expression data rigorously requires taking assumptions into consideration but also relies on using information about network relations that exist among genes. Combining these different elements ...
A comparison of zero-inflated and hurdle models for modeling zero-inflated count data
Counts data with excessive zeros are frequently encountered in practice. For example, the number of health services visits often includes many zeros representing the patients with no utilization during a follo...
A general stochastic model for bivariate episodes driven by a gamma sequence
We propose a new stochastic model describing the joint distribution of ( X , N ), where N is a counting variable while X is the sum of N independent gamma random variables. We present the main properties of this gene...
A flexible multivariate model for high-dimensional correlated count data
We propose a flexible multivariate stochastic model for over-dispersed count data. Our methodology is built upon mixed Poisson random vectors ( Y 1 ,…, Y d ), where the { Y i } are conditionally independent Poisson random...
Generalized fiducial inference on the mean of zero-inflated Poisson and Poisson hurdle models
Zero-inflated and hurdle models are widely applied to count data possessing excess zeros, where they can simultaneously model the process from how the zeros were generated and potentially help mitigate the eff...
Multivariate distributions of correlated binary variables generated by pair-copulas
Correlated binary data are prevalent in a wide range of scientific disciplines, including healthcare and medicine. The generalized estimating equations (GEEs) and the multivariate probit (MP) model are two of ...
On two extensions of the canonical Feller–Spitzer distribution
We introduce two extensions of the canonical Feller–Spitzer distribution from the class of Bessel densities, which comprise two distinct stochastically decreasing one-parameter families of positive absolutely ...
A new trivariate model for stochastic episodes
We study the joint distribution of stochastic events described by ( X , Y , N ), where N has a 1-inflated (or deflated) geometric distribution and X , Y are the sum and the maximum of N exponential random variables. Mod...
A flexible univariate moving average time-series model for dispersed count data
Al-Osh and Alzaid ( 1988 ) consider a Poisson moving average (PMA) model to describe the relation among integer-valued time series data; this model, however, is constrained by the underlying equi-dispersion assumpt...
Spatio-temporal analysis of flood data from South Carolina
To investigate the relationship between flood gage height and precipitation in South Carolina from 2012 to 2016, we built a conditional autoregressive (CAR) model using a Bayesian hierarchical framework. This ...
Affine-transformation invariant clustering models
We develop a cluster process which is invariant with respect to unknown affine transformations of the feature space without knowing the number of clusters in advance. Specifically, our proposed method can iden...
Distributions associated with simultaneous multiple hypothesis testing
We develop the distribution for the number of hypotheses found to be statistically significant using the rule from Simes (Biometrika 73: 751–754, 1986) for controlling the family-wise error rate (FWER). We fin...
New families of bivariate copulas via unit weibull distortion
This paper introduces a new family of bivariate copulas constructed using a unit Weibull distortion. Existing copulas play the role of the base or initial copulas that are transformed or distorted into a new f...
Generalized logistic distribution and its regression model
A new generalized asymmetric logistic distribution is defined. In some cases, existing three parameter distributions provide poor fit to heavy tailed data sets. The proposed new distribution consists of only t...
The spherical-Dirichlet distribution
Today, data mining and gene expressions are at the forefront of modern data analysis. Here we introduce a novel probability distribution that is applicable in these fields. This paper develops the proposed sph...
Item fit statistics for Rasch analysis: can we trust them?
To compare fit statistics for the Rasch model based on estimates of unconditional or conditional response probabilities.
Exact distributions of statistics for making inferences on mixed models under the default covariance structure
At this juncture when mixed models are heavily employed in applications ranging from clinical research to business analytics, the purpose of this article is to extend the exact distributional result of Wald (A...
A new discrete pareto type (IV) model: theory, properties and applications
Discrete analogue of a continuous distribution (especially in the univariate domain) is not new in the literature. The work of discretizing continuous distributions begun with the paper by Nakagawa and Osaki (197...
Density deconvolution for generalized skew-symmetric distributions
The density deconvolution problem is considered for random variables assumed to belong to the generalized skew-symmetric (GSS) family of distributions. The approach is semiparametric in that the symmetric comp...
The unifed distribution
We introduce a new distribution with support on (0,1) called unifed. It can be used as the response distribution for a GLM and it is suitable for data aggregation. We make a comparison to the beta regression. ...
On Burr III Marshal Olkin family: development, properties, characterizations and applications
In this paper, a flexible family of distributions with unimodel, bimodal, increasing, increasing and decreasing, inverted bathtub and modified bathtub hazard rate called Burr III-Marshal Olkin-G (BIIIMO-G) fam...
The linearly decreasing stress Weibull (LDSWeibull): a new Weibull-like distribution
Motivated by an engineering pullout test applied to a steel strip embedded in earth, we show how the resulting linearly decreasing force leads naturally to a new distribution, if the force under constant stress i...
Meta analysis of binary data with excessive zeros in two-arm trials
We present a novel Bayesian approach to random effects meta analysis of binary data with excessive zeros in two-arm trials. We discuss the development of likelihood accounting for excessive zeros, the prior, a...
On ( p 1 ,…, p k )-spherical distributions
The class of ( p 1 ,…, p k )-spherical probability laws and a method of simulating random vectors following such distributions are introduced using a new stochastic vector representation. A dynamic geometric disintegra...
A new class of survival distribution for degradation processes subject to shocks
Many systems experience gradual degradation while simultaneously being exposed to a stream of random shocks of varying magnitudes that eventually cause failure when a shock exceeds the residual strength of the...
A new extended normal regression model: simulations and applications
Various applications in natural science require models more accurate than well-known distributions. In this context, several generators of distributions have been recently proposed. We introduce a new four-par...
Multiclass analysis and prediction with network structured covariates
Technological advances associated with data acquisition are leading to the production of complex structured data sets. The recent development on classification with multiclass responses makes it possible to in...
High-dimensional star-shaped distributions
Stochastic representations of star-shaped distributed random vectors having heavy or light tail density generating function g are studied for increasing dimensions along with corresponding geometric measure repre...
A unified complex noncentral Wishart type distribution inspired by massive MIMO systems
The eigenvalue distributions from a complex noncentral Wishart matrix S = X H X has been the subject of interest in various real world applications, where X is assumed to be complex matrix variate normally distribute...
Particle swarm based algorithms for finding locally and Bayesian D -optimal designs
When a model-based approach is appropriate, an optimal design can guide how to collect data judiciously for making reliable inference at minimal cost. However, finding optimal designs for a statistical model w...
Admissible Bernoulli correlations
A multivariate symmetric Bernoulli distribution has marginals that are uniform over the pair {0,1}. Consider the problem of sampling from this distribution given a prescribed correlation between each pair of v...
On p -generalized elliptical random processes
We introduce rank- k -continuous axis-aligned p -generalized elliptically contoured distributions and study their properties such as stochastic representations, moments, and density-like representations. Applying th...
Parameters of stochastic models for electroencephalogram data as biomarkers for child’s neurodevelopment after cerebral malaria
The objective of this study was to test statistical features from the electroencephalogram (EEG) recordings as predictors of neurodevelopment and cognition of Ugandan children after coma due to cerebral malari...
A new generalization of generalized half-normal distribution: properties and regression models
In this paper, a new extension of the generalized half-normal distribution is introduced and studied. We assess the performance of the maximum likelihood estimators of the parameters of the new distribution vi...
Analytical properties of generalized Gaussian distributions
The family of Generalized Gaussian (GG) distributions has received considerable attention from the engineering community, due to the flexible parametric form of its probability density function, in modeling ma...
A new Weibull- X family of distributions: properties, characterizations and applications
We propose a new family of univariate distributions generated from the Weibull random variable, called a new Weibull-X family of distributions. Two special sub-models of the proposed family are presented and t...
The transmuted geometric-quadratic hazard rate distribution: development, properties, characterizations and applications
We propose a five parameter transmuted geometric quadratic hazard rate (TG-QHR) distribution derived from mixture of quadratic hazard rate (QHR), geometric and transmuted distributions via the application of t...
A nonparametric approach for quantile regression
Quantile regression estimates conditional quantiles and has wide applications in the real world. Estimating high conditional quantiles is an important problem. The regular quantile regression (QR) method often...
Mean and variance of ratios of proportions from categories of a multinomial distribution
Ratio distribution is a probability distribution representing the ratio of two random variables, each usually having a known distribution. Currently, there are results when the random variables in the ratio fo...
The power-Cauchy negative-binomial: properties and regression
We propose and study a new compounded model to extend the half-Cauchy and power-Cauchy distributions, which offers more flexibility in modeling lifetime data. The proposed model is analytically tractable and c...
Families of distributions arising from the quantile of generalized lambda distribution
In this paper, the class of T-R { generalized lambda } families of distributions based on the quantile of generalized lambda distribution has been proposed using the T-R { Y } framework. In the development of the T - R {
Risk ratios and Scanlan’s HRX
Risk ratios are distribution function tail ratios and are widely used in health disparities research. Let A and D denote advantaged and disadvantaged populations with cdfs F ...
Joint distribution of k -tuple statistics in zero-one sequences of Markov-dependent trials
We consider a sequence of n , n ≥3, zero (0) - one (1) Markov-dependent trials. We focus on k -tuples of 1s; i.e. runs of 1s of length at least equal to a fixed integer number k , 1≤ k ≤ n . The statistics denoting the n...
Quantile regression for overdispersed count data: a hierarchical method
Generalized Poisson regression is commonly applied to overdispersed count data, and focused on modelling the conditional mean of the response. However, conditional mean regression models may be sensitive to re...
Describing the Flexibility of the Generalized Gamma and Related Distributions
The generalized gamma (GG) distribution is a widely used, flexible tool for parametric survival analysis. Many alternatives and extensions to this family have been proposed. This paper characterizes the flexib...
- ISSN: 2195-5832 (electronic)
- Search Menu
- Sign in through your institution
- Advance Articles
- Author Guidelines
- Submission Site
- Open Access Policy
- Self-Archiving Policy
- Why publish with Series A?
- About the Journal of the Royal Statistical Society Series A: Statistics in Society
- About The Royal Statistical Society
- Editorial Board
- Advertising & Corporate Services
- Journals on Oxford Academic
- Books on Oxford Academic

B. De Stavola and M. Elliott Editorial Board
About the journal
The Journal of the Royal Statistical Society, Series A (Statistics in Society) , publishes papers that demonstrate how statistical thinking, design and analyses play a vital role in life and benefit society.

Author resources
Learn about how to submit your article, our publishing process, and tips on how to promote your article.
Find out more

Submit Your Research
Explore the author guidelines and submit your research.
Access the guidelines

High Impact Research Collection
Explore papers from the Journal of the Royal Statistical Society: Series A with high citations.

Email alerts
Register for email alerts to stay up to date with the Royal Statistical Society journals.
Latest articles
Latest posts on x.

Discussion Papers
Explore past discussion papers from the Journal of the Royal Statistical Society: Series A .

Special Collections
- Special issues
- Virtual issues

Journal webinars are held every few months and last about an hour. Learn more about upcoming webinars.

About the Royal Statistical Society
Learn more about the RSS and explore the benefits of membership.

- Accessibility
Oxford University Press is committed to making its products accessible to and inclusive of all our users, including those with visual, hearing, cognitive, or motor impairments.
Find out more in our accessibility statement

Committee on Publication Ethics (COPE)
This journal is a member of and subscribes to the principles of the Committee on Publication Ethics (COPE)
publicationethics.org

Read and Publish deals
Authors interested in publishing in Journal of the Royal Statistical Society Series A: Statistics in Society may be able to publish their paper Open Access using funds available through their institution’s agreement with OUP.
Find out if your institution is participating

Coming soon: RSS: Data Science
A new fully Open Access journal in the RSS family, RSS: Data Science will welcome submissions from across the breadth of data science, including AI, statistics, machine learning, econometrics, bioinformatics, and beyond.
Related Titles

- Recommend to Your Librarian
- Advertising & Corporate Services
- Journals Career Network
- Email Alerts
Affiliations
- Online ISSN 1467-985X
- Print ISSN 0964-1998
- Copyright © 2024 Royal Statistical Society
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

IMAGES
VIDEO
COMMENTS
Introduction. Statistical analysis is necessary for any research project seeking to make quantitative conclusions. The following is a primer for research-based statistical analysis. It is intended to be a high-level overview of appropriate statistical testing, while not diving too deep into any specific methodology.
1 Introduction. Statistics is a set of methods used to analyze data. The statistic is present in all areas of science involving the. collection, handling and sorting of data, given the insight of ...
Table of contents. Step 1: Write your hypotheses and plan your research design. Step 2: Collect data from a sample. Step 3: Summarize your data with descriptive statistics. Step 4: Test hypotheses or make estimates with inferential statistics.
Overview. Statistical Papers is a forum for presentation and critical assessment of statistical methods encouraging the discussion of methodological foundations and potential applications. The Journal stresses statistical methods that have broad applications, giving special attention to those relevant to the economic and social sciences.
Provides well-organized coverage of statistical analysis and applications in biology, kinesiology, and physical anthropology with comprehensive insights into the techniques and interpretations of R, SPSS®, Excel®, and Numbers® output An Introduction to Statistical Analysis in Research: With Applications in the Biological and Life Sciences develops a conceptual foundation in statistical ...
Hypothesis testing is a formal process of statistical analysis using inferential statistics. The goal of hypothesis testing is to compare populations or assess relationships between variables using samples. Hypotheses, or predictions, are tested using statistical tests. Statistical tests also estimate sampling errors so that valid inferences ...
The purpose of statistical analysis is to clarify and not confuse. It is a tool for answering questions. It allows us to take large bodies of information and summarize them with a few simple statements. It lets us come to solid conclusions even when the realities of the research world make it difficult to isolate the problems we seek to study.
RESEARCH ARTICLE. Prior effective sample size for exponential family distributions with multiple parameters. Ryota Tamanoi, First Published: 9 May 2024; Abstract; ... Statistical Analysis and Data Mining. Related Titles Issue Volume 14, Issue 3. May/June 2024. Issue Volume 92, Issue 1. Pages: 1-135. April 2024.
A support vector machine based drought index for regional drought analysis. Mohammed A Alshahrani. , Muhammad Laiq. & Muhammad Nabi. Article. 25 April 2024 | Open Access.
This article is a practical guide to conducting data analysis in general literature reviews. The general literature review is a synthesis and analysis of published research on a relevant clinical issue, and is a common format for academic theses at the bachelor's and master's levels in nursing, physiotherapy, occupational therapy, public health and other related fields.
The knowledge of statistics is usually quite limited among researchers representing the field of life sciences, particularly when it comes to constraints imposed on the use of statistical tools and possible interpretations. A common mistake is that researchers take for granted the ability to perform a valid statistical analysis.
Measuring variables. When planning a research design, you should operationalise your variables and decide exactly how you will measure them.. For statistical analysis, it's important to consider the level of measurement of your variables, which tells you what kind of data they contain:. Categorical data represents groupings. These may be nominal (e.g., gender) or ordinal (e.g. level of ...
Statistical analysis involves gathering and testing quantitative data to make inferences about the world. ... In the methods section of an APA research paper, you report in detail the participants, measures, and procedure of your study. 264. APA format for academic papers and essays
for statistical data analysis without additional costs. With the help of the R system for statistical computing, research really becomes reproducible when both the data and the results of all data analysis steps reported in a paper are available to the readers through an Rtranscript file. Ris most widely used for
important writing you will do for the paper. IMHO your reader will either be interested and continuing on with your paper, or... A scholarly introduction is respectful of the literature. In my experience, the introduction is part of a paper that I will outline relatively early in the process, but will nish and repeatedly edit at the end of the ...
In this paper, we have provided a simple statistical analysis of the novel Coronavirus (COVID-19) outbreak in Italy and Spain—two of the worst affected countries in Europe. ... National Bureau of Economic Research, Working Paper 26867. 14. Wang P., Zheng X., Li J. and Zhu B., 2020. Prediction of epidemic trends in COVID-19 with logistic model ...
Our methodology is built upon mixed Poisson random vectors ( Y1 ,…, Yd ), where the { Yi } are conditionally independent Poisson random... Alexander D. Knudson, Tomasz J. Kozubowski, Anna K. Panorska and A. Grant Schissler. Journal of Statistical Distributions and Applications 2021 8 :6. Research Published on: 16 March 2021.
The data analysis report isn't quite like a research paper or term paper in a class, nor like aresearch article in a journal. It is meant, primarily, to start an organized conversation between you and your client/collaborator. In that sense it is a kind of "internal" communication, sort o f like an extended memo. On the other hand it
About the journal. The Journal of the Royal Statistical Society, Series A (Statistics in Society), publishes papers that demonstrate how statistical thinking, design and analyses play a vital role in life and benefit society. Learn more.
The photometric characteristics of the input and enhanced images have been statistically characterized, and the contrast enhancement assessed by a state-of-the-art metric. The statistical analysis of the text colour coordinates was carried out to develop supervised and unsupervised image segmentation, isolating the text.
ANOVA and MANOVA tests are used when comparing the means of more than two groups (e.g., the average heights of children, teenagers, and adults). Predictor variable. Outcome variable. Research question example. Paired t-test. Categorical. 1 predictor. Quantitative. groups come from the same population.
Abstract. The Yale Law and Political Economy ("LPE") Project began in 2017 following the surprising election of Donald Trump as President. In that time, LPE has increasingly emerged into an intellectual and ideological movement particularly at elite law schools involving the efforts of numerous leading academics, substantial foundation backing, and its own dedicated journal.
There are 3 main types of descriptive statistics: The distribution concerns the frequency of each value. The central tendency concerns the averages of the values. The variability or dispersion concerns how spread out the values are. You can apply these to assess only one variable at a time, in univariate analysis, or to compare two or more, in ...