- Skip to main content
- Keyboard shortcuts for audio player
- Your Health
- Treatments & Tests
- Health Inc.
- Public Health

The CRISPR Revolution
He inherited a devastating disease. a crispr gene-editing breakthrough stopped it.

Patrick Doherty volunteered for a new medical intervention of gene-editor infusions for the treatment of genetically-based diseases. Patrick Doherty hide caption
Patrick Doherty volunteered for a new medical intervention of gene-editor infusions for the treatment of genetically-based diseases.
Patrick Doherty had always been very active. He trekked the Himalayas and hiked trails in Spain.
But about a year and a half ago, he noticed pins and needles in his fingers and toes. His feet got cold. And then he started getting out of breath any time he walked his dog up the hills of County Donegal in Ireland where he lives.
"I noticed on some of the larger hill climbs I was getting a bit breathless," says Doherty, 65. "So I realized something was wrong."
Doherty found out he had a rare, but devastating inherited disease — known as transthyretin amyloidosis — that had killed his father. A misshapen protein was building up in his body, destroying important tissues, such as nerves in his hands and feet and his heart.
Doherty had watched others get crippled and die difficult deaths from amyloidosis.
"It's terrible prognosis," Doherty says. "This is a condition that deteriorates very rapidly. It's just dreadful."
So Doherty was thrilled when he found out that doctors were testing a new way to try to treat amyloidosis. The approach used a revolutionary gene-editing technique called CRISPR , which allows scientists to make very precise changes in DNA.
"I thought: Fantastic. I jumped at the opportunity," Doherty says.
On Saturday, researchers reported the first data indicating that the experimental treatment worked, causing levels of the destructive protein to plummet in Doherty's body and the bodies of five other patients treated with the approach.
"I feel fantastic," Doherty says. "It's just phenomenal."
The advance is being hailed not just for amyloidosis patients but also as a proof-of-concept that CRISPR could be used to treat many other, much more common diseases. It's a new way of using the innovative technology.
"This is a major milestone for patients," says Jennifer Doudna of the University of California, Berkeley, who shared a Nobel Prize for her work helping develop CRISPR.
"While these are early data, they show us that we can overcome one of the biggest challenges with applying CRISPR clinically so far, which is being able to deliver it systemically and get it to the right place," Doudna says.
CRISPR has already been shown to help patients suffering from the devastating blood disorders sickle cell disease and beta thalassemia . And doctors are trying to use it to treat cancer and to restore vision to people blinded by a rare genetic disorder.
But those experiments involve taking cells out of the body, editing them in the lab, and infusing them back in or injecting CRISPR directly into cells that need fixing.
The study Doherty volunteered for is the first in which doctors are simply infusing the gene-editor directly into patients and letting it find its own way to the right gene in the right cells. In this case, it's cells in the liver making the destructive protein.
"This is the first example in which CRISPR-Cas9 is injected directly into the bloodstream — in other words systemic administration — where we use it as a way to reach a tissue that's far away from the site of injection and very specifically use it to edit disease-causing genes," says John Leonard, the CEO of Intellia Therapeutics , which is sponsoring the study.
Doctors infused billions of microscopic structures known as nanoparticles carrying genetic instructions for the CRISPR gene-editor into four patients in London and two in New Zealand. The nanoparticles were absorbed by their livers, where they unleashed armies of CRISPR gene-editors. The CRISPR editor homed in on the target gene in the liver and sliced it, disabling production of the destructive protein.
Within weeks, the levels of protein causing the disease plummeted, especially in the volunteers who received a higher dose. Researchers reported at the Peripheral Nerve Society Annual Meeting and in a paper published in The New England Journal of Medicine .
"It really is exciting," says Dr. Julian Gillmore , who is leading the study at the University College London, Royal Free Hospital.
"This has the potential to completely revolutionize the outcome for these patients who have lived with this disease in their family for many generations. It's decimated some families that I've been looking after. So this is amazing," Gillmore says.
The patients will have to be followed longer, and more patients will have to be treated, to make sure the treatment's safe, and determine how much it's helping, Gillmore stresses. But the approach could help those struck by amyloidosis that isn't inherited, which is a far more common version of the disease, he says.
Moreover, the promising results potentially open the door for using the same approach to treatment of many other, more common diseases for which taking cells out of the body or directly injecting CRISPR isn't realistic, including heart disease, muscular dystrophy and brain diseases such as Alzheimer's.
"This is really opening a new era as we think about gene-editing where we can begin to think about accessing all kinds of different tissue in the body via systemic administration," Leonard says.
Other scientists who are not involved in the research agree.
"This is a wonderful day for the future of gene-editing as a medicine," agree Fyodor Urnov , a professor of genetics at the University of California, Berkeley. "We as a species are watching this remarkable new show called: our gene-edited future."
Doherty says he started feeling better within weeks of the treatment and has continued to improve in the weeks since then.
"I definitely feel better," he told NPR. "I'm speaking to you from upstairs in our house. I climbed stairs to get up here. I would have been feeling breathless. I'm thrilled."
Subscribe or renew today
Every print subscription comes with full digital access
Science News
Here’s why some pigeons do backflips.
Meet the scientist homing in on the genes involved in making parlor roller pigeons do backward somersaults.
A genetic parasite may explain why humans and other apes lack tails
Ancient viruses helped speedy nerves evolve, more stories in genetics.

Newfound immune cells are responsible for long-lasting allergies
A specialized type of immune cell appears primed to make the type of antibodies that lead to allergies, two research groups report.

Geneticist Krystal Tsosie advocates for Indigenous data sovereignty
A member of the Navajo Nation, she believes Indigenous geneticists have a big role to play in protecting and studying their own data.

How ancient herders rewrote northern Europeans’ genetic story
New DNA analyses show the extent of the Yamnaya people’s genetic reach starting 5,000 years ago and how it made descendants prone to diseases like MS.

Fetuses make a protein that causes morning sickness in pregnancy
A hormone called GDF15 triggers a part of the brain involved in nausea and vomiting, a new study finds. Blocking its action may lead to treatments.

Why Huntington’s disease may take so long to develop
Repeated bits of the disease-causing gene pile up in some brain cells. New treatments could involve stopping the additions.

Here’s how high-speed diving kingfishers may avoid concussions
Understanding the genetic adaptations that protect the birds’ brains when they dive for food might one day offer clues to protecting human brains.

These 8 GMOs tell a brief history of genetic modification
Since the first genetically modified organism 50 years ago, GMOs have brought us disease-resistant crops, new drugs and more.

Most of today’s gene therapies rely on viruses — and that’s a problem
The next big strides in gene therapy for rare diseases may come from CRISPR and new approaches to delivery.

In a first, genetically modified silkworms produced pure spider silk
An effort to engineer silkworms to produce spider silk brings us closer than ever to exploiting the extraordinary properties of this arachnid fiber.
Subscribers, enter your e-mail address for full access to the Science News archives and digital editions.
Not a subscriber? Become one now .
- U.S. Department of Health & Human Services

- Virtual Tour
- Staff Directory
- En Español
You are here
News releases.
News Release
Thursday, January 21, 2021
NIH researchers identify new genetic disorder that affects brain, craniofacial skeleton
Analysis of patients with rare condition uncovers key pathway for human development.

Researchers at the National Institutes of Health have discovered a new genetic disorder characterized by developmental delays and malformations of the brain, heart and facial features. Named linkage-specific-deubiquitylation-deficiency-induced embryonic defects syndrome (LINKED), it is caused by a mutated version of the OTUD5 gene, which interferes with key molecular steps in embryo development. The findings indicate that the newly identified pathway may be essential for human development and may also underlie other disorders that are present at birth. The information will help scientists better understand such diseases — both common and rare — and improve patient care. The results were reported Jan. 20, 2021 in Science Advances .
“Our discovery of the dysregulated neurodevelopmental pathway that underlies LINKED syndrome was only possible through the teamwork of geneticists, developmental biologists and biochemists from NIH,” said Achim Werner, Ph.D., an investigator at the National Institute of Dental and Craniofacial Research (NIDCR) and lead author. “This collaboration provided the opportunity to pinpoint the likely genetic cause of disease, and then take it a step further to precisely define the sequence of cellular events that are disrupted to cause the disease.”
The project began when David B. Beck, M.D., Ph.D., a clinical fellow in the laboratory of Dan Kastner M.D., Ph.D., at the National Human Genome Research Institute (NHGRI) and co-first author, was asked to consult on a male infant who had been born with severe birth defects that included abnormalities of the brain, craniofacial skeleton, heart and urinary tract. An in-depth examination of siblings’ and family members’ genomes, combined with genetic bioinformatics analyses, revealed a mutation in the OTUD5 gene as the likely cause of the condition. Through outreach to other researchers working on similar problems, Beck found seven additional males ranging from 1 to 14 years of age who shared symptoms with the first patient and had varying mutations in the OTUD5 gene.
The gene contains instructions for making the OTUD5 enzyme, which is involved in ubiquitylation, a process that molecularly alters a protein to change its function. Ubiquitylation plays a role in governing cell fate, where stem cells are instructed to become specific cell types in the early stages of embryo development.
“Based on the genetic evidence, I was pretty sure OTUD5 mutations caused the disease, but I didn’t understand how this enzyme, when mutated, led to the symptoms seen in our patients,” said Beck. “For this reason we sought to work with Dr. Werner’s group, which specializes in using biochemistry to understand the functions of enzymes like OTUD5.”
To start, the NIH team examined cells taken from patient samples, which were processed at the NIH Clinical Center. Normally, OTUD5 edits or removes molecular tags on certain proteins (substrates) to regulate their function. But in cells from patients with OTUD5 mutations, this activity was impaired.
Using a method to return mature human cells to the stem cell-like state of embryo cells, the scientists found that OTUD5 mutations were linked to abnormalities in the development of neural crest cells, which give rise to tissues of the craniofacial skeleton, and of neural precursors, cells that eventually give rise to the brain and spinal cord.
In further experiments, the team discovered that the OTUD5 enzyme acts on a handful of protein substrates called chromatin remodelers. This class of proteins physically alters the tightly packed strands of DNA in a cell’s nucleus to make certain genes more accessible for being turned on, or expressed.
With help from collaborators led by Pedro Rocha Ph.D., an investigator at the National Institute of Child Health and Human Development (NICHD), the team found that chromatin remodelers targeted by OTUD5 help enhance expression of genes that control the cell fate of neural precursors during embryo development.
Taken together, the researchers concluded, OTUD5 normally keeps these chromatin remodelers from being tagged for destruction. But when OTUD5 is mutated, its protective function is lost and the chromatin remodelers are destroyed, leading to abnormal development of neural precursors and neural crest cells. Ultimately, these changes can lead to some of the birth defects seen in LINKED patients.
“Several of the chromatin remodelers OTUD5 interacts with are mutated in Coffin Siris and Cornelia de Lange syndromes, which have clinically overlapping features with LINKED syndrome,” said Werner. “This suggests that the mechanism we discovered is part of a common developmental pathway that, when mutated at various points, will lead to a spectrum of disease.”
“We were surprised to find that OTUD5 elicits its effects through multiple, functionally related substrates, which reveals a new principle of cellular signaling during early embryonic development,” said Mohammed A. Basar, Ph.D., a postdoctoral fellow in Werner’s lab and co-first author of the study. “These findings lead us to believe that OTUD5 may have far-reaching effects beyond those identified in LINKED patients.”
In future work, Werner’s team plans to more fully investigate the role that OTUD5 and similar enzymes play in development. The researchers hope the study can serve as a guiding framework for unraveling the causes of other undiagnosed diseases, ultimately helping clinicians better assess and care for patients.
“We’re finally able to provide families with a diagnosis, bringing an end to what is often a long and exhausting search for answers,” said Beck.
This research was supported by NIH intramural research programs at NIDCR, NICHD, NHGRI and the NIH Undiagnosed Diseases Program. Support also came from the Estonian Research Council, Japan Society for the Promotion of Science and the Japan Agency for Medical Research and Development.
About the National Institute of Dental and Craniofacial Research: NIDCR is the nation’s leading funder of research on oral, dental and craniofacial health.
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit www.nih.gov .
NIH…Turning Discovery Into Health ®
Beck D.B., Basar M.A., et al. Linkage-specific deubiquitylation by OTUD5 defines an embryonic pathway intolerant to genomic variation . Science Advances. Jan 21, 2021. DOI: 10.1126/sciadv.abe2116
Connect with Us
- More Social Media from NIH

Transforming the understanding and treatment of mental illnesses.
Información en español
Celebrating 75 Years! Learn More >>
- Science News
- Meetings and Events
- Social Media
- Press Resources
- Email Updates
- Innovation Speaker Series
Researchers Unlock Genetic Mutations Contributing to Disorders in the Brain
April 10, 2023 • Research Highlight
Epilepsies are chronic neurological disorders in which large groups of neurons firing at the same time generate electrical activity that causes seizures and involuntary movements. They are one of the most common brain diseases in children and, in almost a quarter of cases, patients do not respond to standard medical treatments. Life-threatening treatment-resistant epilepsy often results from tissue that was damaged or developed abnormally during prenatal brain formation, known as malformations of cortical development (MCD).
Epilepsy resulting from MCD is a rare but serious condition. Although some types of epilepsy run in families, the genetic cause of MCD is unclear. New research funded by the National Institute of Mental Health (NIMH), National Institute of Neurological Disorders and Stroke, and National Institute on Aging sheds light on genetic mutations that may play a key role in the development of epilepsies. The study provides insights that could lead to improved diagnosis and treatment of diseases with origins in early brain development.
Led by Joseph Gleeson, M.D. , at the University of California San Diego and the Rady Children’s Institute for Genomic Medicine, the study was a multicenter international collaboration. The researchers looked for mutations in the brain that may contribute to MCD. They performed genetic profiling of tissue using advanced detection techniques and best practice guidelines from the Brain Somatic Mosaicism Network —an NIMH-supported network of investigative teams working together to study mutations present in a small subset of brain cells.
Almost 300 children with diverse forms of MCD provided brain tissue through the Focal Cortical Dysplasia Neurogenetics Consortium . Brain samples were collected as part of surgery to treat epilepsy. For each person, paired blood or saliva samples were also collected, as were parental samples when available. The researchers included brain tissue from a small sample of people without neurological conditions for comparison and validated a subset of identified genes via patient biopsies and in mice.
Comprehensive screening to identify genetic causes of MCD proceeded in three phases:
- Targeted examination of genes in the mTOR pathway, which regulates cell growth, proliferation, and metabolism and shows excessive signaling in the brains of people with epilepsy
- Unbiased gene discovery to identify new genes that may be associated with MCD
- Independent testing in a new sample to confirm the genes identified in the first two phases
Additional analyses looked for networks of genes with related functions involved in brain development and at links between identified genes and clinical and behavioral features of the disease.
This study identified 69 mutated genes associated with MCD. Of these, 60 were genes linked to MCD for the first time. Twelve of the mutated genes were recurrently mutated, meaning they were identified in at least two different patient brain samples, giving more confidence that they contribute to MCD. Among the recurrently mutated genes were two genes linked to MCD for the first time and another three genes identified in prior studies. These data suggest that researchers have only scratched the surface of the number of genes involved in epilepsy and may identify more genes in future studies.
The results also confirmed the critical role of the mTOR pathway. This pathway is dysregulated in several human diseases, including cancer and diabetes. As such, the mutations could have implications for risk for any number of diseases and disorders.
To test the function of the mutations, the researchers introduced mutated or non-mutated forms of the identified MCD genes into a small region of the brain in developing mice. Introduction of the mutated genes led to the development of brain abnormalities similar to those seen in humans with MCD, indicating that many of the mutated genes likely contribute to features of the disease. Further analyses revealed four major networks into which the mutated genes clustered, all of which play critical roles during early brain development. These groups of genes correlated with clinical features of the disease. Together, the results showed that the mutated genes are vital to cortical development and related to patient outcomes later in life.
The findings of this study have important implications for treatment-resistant epilepsy and related diseases, as well as for human brain development. The identified genes could offer potential drug targets, help inform new clinical classifications and diagnoses, and ultimately lead to personalized treatments or early interventions for a range of mental and physical health conditions.
The current sample size was larger than in previous studies, leading to the discovery of many new genes. The researchers’ use of state-of-the-art methods and independent validation of genes also enhanced confidence in the results. However, confirming the current set of genes and identifying new MCD-related genes will require replication in larger samples. Future research taking advantage of this study’s innovative roadmap for studying rare genetic variants will also help answer important questions, such as the contribution of environmental versus genetic factors in disease.
Chung, C., Yang, X., Bae, T., Vong, K. I., Mittal, S., Donkels, C., Phillips, H. W., Li, Z., Marsh, A. P. L., Breuss, M. W., Ball, L. L., Garcia, C. A. B., Gu, J., Xu, M., Barrows, C., James, K. N., Stanley, V., Nidhiry, A. S., Khoury, S. … Gleeson, J. G. (2023). Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development. Nature Genetics , 55 , 209 – 220. https://doi.org/10.1038/s41588-022-01276-9
MH108898 , MH124890 , AG070462 , NS083823

New research into genetic mutations may pave the way for more effective gene therapies
Assistant Professor, Biology, University of Toronto
Disclosure statement
Alex Nguyen Ba receives funding from the Natural Sciences and Engineering Research Council of Canada.
University of Toronto provides funding as a founding partner of The Conversation CA.
University of Toronto provides funding as a member of The Conversation CA-FR.
View all partners
Consider a living cell, which can have thousands of genes. Now think of these genes as dials that can be tweaked to change how the cell grows in a given environment. Tweaking a gene can either increase or decrease growth, and this is made more complex considering these dials are interconnected with each other, like cogs in a machine.
While scientists are now able to edit genes in laboratory conditions and attempt to produce findings that may lead to cures, evolution has been doing this for billions of years. Evolution is the natural process that turns these dials, allowing populations to adapt. However, unlike scientists, evolution turns these dials randomly as mutations affect the function of genes.
One underlying hypothesis in evolutionary theory — the evolutionary contingency hypothesis — has been that this tuning can have chaotic behaviours. Or, in other words, dials tweaked early in the process can dramatically alter later evolutionary potential.
Stephen Jay Gould was a famous proponent of this theory, arguing in his 1989 book Wonderful Life that since beneficial mutations occur randomly, chance must play an important role in evolutionary diversification.
Read more: Does our DNA really determine our intelligence and health?
If this hypothesis is true, it affects how scientists should edit genes in the laboratory as they will face the chaotic interconnections of our cells. Our work set out to test this hypothesis.
Resolving an evolutionary paradox
We can observe the process of evolution in the laboratory under extremely well-controlled conditions. We have done so by growing populations of micro-organisms for hundreds — even thousands — of days .
Since these organisms divide and reproduce so quickly, this process represents thousands of generations of growth. These experiments have allowed us to pinpoint precisely when , and how, beneficial mutations co-occur and compete to take over the population.
One striking observation from every single one of these experiments is that increases in fitness slow down over time at a rate that is surprisingly reproducible. Despite accumulating different mutations, different populations show remarkably predictable diminishing returns in how fast they adapt.
In contrast with the seemingly chaotic behaviour of mutations, fitness or growth changes are highly predictable. This has led many to hypothesize that this order of mutation is an inherent consequence of the way biological systems have evolved.
This striking hypothesis is at odds with the idea that the specifics of an organism’s biology matter for evolution . In other words, it has been difficult to prove that the order in which evolution turns dials has any impact on the future.
The answer to the paradox
My team was able to show that the answer to resolving this paradox lies within the interconnected gene network of the cell itself.
For evolution to work, the dial-tuning must be precise: even if the net outcome is beneficial, adjusting one set of linked dials can trickle down and affect other previously correctly placed dials. As evolution continues, the probability of breaking harmoniously-tuned dials grows. This seemingly simple principle explains why the rate of evolutionary improvements typically slows down over time.

Resolving this paradox experimentally was not an easy task. After all, how can one show the entanglement of dials within the cell? In our recent study , we tackled this challenge by systematically trying out every possible combination of 10 key beneficial mutations and looking at how they affect the growth of cells.
Read more: Human genome editing offers tantalizing possibilities – but without clear guidelines, many ethical questions still remain
By testing out combinations of mutations, we were able to reliably understand which mutations were entangled together (this entanglement is known as epistasis) and for just 10 mutations, over 1,000 combinations had to be generated.
How this affects genetic precision medicine
Current futuristic technologies tout the ability to generate precise single mutations within our own genomes with the hope that this can be used to repair non-functional genetic variants. For example, prime editing is an effective “search-and-replace” genome editing technology.
One important concern with these approaches is they can introduce undesired mutations at the same time. However, even as scientists solve these concerns, the field of human genetics has often overlooked the importance of the interconnectedness of genes .
Our study demonstrates that bioengineers should think not only about the effect a mutation has on the gene it is in, but also about the effect of the mutation in the context of all other variations in our genomes. Altering the function of any of our genes can affect our interconnected cellular networks.
This is compounded by the fact that all of us carry hundreds of extremely rare variants, which means each of us carries a unique interconnected network of genes. These personalized networks make us who we are.
Read more: Somatic genome editing therapies are becoming a reality – but debate over ethics, equitable access and governance continue
Genome interpretation is at the heart of genetic testing for disease. And while scientists have made some progress in identifying key pathogenic genetic variants (those that can cause disease), our findings demonstrate that classifying a variant as pathogenic or benign requires us to also understand how the other genetic dials in our cells are tuned.
- Bioengineering
- Human genome
- Human genetics
- Genetic engineering
- survival of the fittest
- Gene mutation

Events Officer

Lecturer (Hindi-Urdu)

Director, Defence and Security

Opportunities with the new CIEHF

School of Social Sciences – Public Policy and International Relations opportunities
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Portland Press Opt2Pay

Recent developments in genetic/genomic medicine
Rachel h. horton.
Clinical Ethics and Law, Faculty of Medicine, University of Southampton, Southampton, United Kingdom
Anneke M. Lucassen
Advances in genetic technology are having a major impact in the clinic, and mean that many perceptions of the role and scope of genetic testing are having to change. Genomic testing brings with it a greater opportunity for diagnosis, or predictions of future diagnoses, but also an increased chance of uncertain or unexpected findings, many of which may have impacts for multiple members of a person’s family. In the past, genetic testing was rarely able to provide rapid results, but the increasing speed and availability of genomic testing is changing this, meaning that genomic information is increasingly influencing decisions around patient care in the acute inpatient setting. The landscape of treatment options for genetic conditions is shifting, which has evolving implications for clinical discussions around previously untreatable disorders. Furthermore, the point of access to testing is changing with increasing provision direct to the consumer outside the formal healthcare setting. This review outlines the ways in which genetic medicine is developing in light of technological advances.
Introduction
The past two decades have seen major shifts in our technical ability to sequence genetic information at scale. Historically, genetic testing tended to consist of either highly detailed molecular testing of nominated single genes, or broad genome-wide dosage screening at low resolution, for example karyotyping [ 1 , 2 ]. Genome sequencing was too slow and too expensive to be used in clinical contexts: for example the Human Genome Project, which was 99% complete in 2004, cost three billion dollars and took 13 years to sequence [ 3 ].
More recently, advances in sequencing technology have made it possible to undertake broad genetic testing on an individual patient basis within a clinically useful timeframe, via exome and genome sequencing. Exome tests sequence the entire protein-coding region of the genome, representing less than 2% of the genome but containing approximately 85% of known disease-causing variants [ 4 ]; genome sequencing encompasses the exome but also sequences all the non-protein-coding DNA. Initially implementation of such tests was via clinical research studies such as the Deciphering Developmental Disorders project [ 5 ], but more recently exome sequencing has been utilised as a clinical diagnostic test [ 6 ]. Genome sequencing is also due to transition to being available as a standard NHS test in June 2019, having previously only been available via initiatives such as the 100,000 Genomes Project [ 7 ].
Sequencing technology has improved in depth as well as breadth, and this has been of importance in better understanding cancer. The ability to sequence cancer genomes has led to rapid identification of driver mutations and has helped to work out the complex relationships between different cancer subclones over space and time, demonstrating the enormous heterogeneity of cancers and the difficulty of successfully treating them [ 8 ]. As sequencing techniques have advanced to the level where tiny amounts of tumour or individual cells can be sequenced, it has been possible to identify previously unknown mutational mechanisms, such as chromothripsis 1 [ 9 ] and kataegis 2 [ 10 ].
However, our ability to generate genomic data has substantially outstripped our ability to interpret its significance for an individual, and while improvements in genomic technology are in many cases driving improvements in healthcare, we are also encountering new problems as genomic testing shifts into the clinical setting. The Global Alliance for Genomics and Health (GA4GH) predicts that by 2025, over 60 million people will have had their genome sequenced in a healthcare context [ 11 ], but pathways for managing the output from genome sequencing are still in their infancy. The detailed but unfocused approach of genomic tests gives opportunities to answer questions that go beyond the problems that led to a patient having a test. However, deciding which of the multitude of possible outputs from genomic tests should be considered a ‘result’ at any given time is very challenging, not least because the links between many genetic variants and diseases are often unproven or poorly understood [ 12 ]. Multidisciplinary input and collaboration are increasingly key to interpreting the significance of genomic results. This review discusses the developments in practice that are evolving as a result of increasing use of genomic technologies.
New disease gene discovery and changing concepts of diagnosis
Exome and genome sequencing are powerful diagnostic tools – for example the Deciphering Developmental Disorders project, which recruited patients with severe undiagnosed disorders (who had generally already had any currently available diagnostic genetic testing), achieved a 40% diagnosis rate via trio exome sequencing for the first 1133 family trios in the study [ 13 ]. The search for a diagnosis has often been described as a journey [ 14 ], with parents of children with rare genetic disorders anticipating that a diagnosis may guide treatment, prognosis, acceptance and social support [ 15 ]. However, identification of new rare disease genes may be changing the impact of receiving a diagnosis, and in many cases very little is known about the long-term effects of newly identified genetic conditions.
Historically when making a genetic diagnosis, it has usually been possible to give families some information regarding prognosis, and to provide some parameters as to what to expect for the future, based on previous experience of what has happened for other children affected by the same condition. Now, while in some situations due to strong phenotypic match it is possible to be confident that a child’s rare disease has been caused by pathogenic variants in a recently described rare disease gene, often this provides little information about a child’s future.
We are increasingly in the position of learning about the effects of possible disease-causing variation(s) in a gene through meeting the patients in whom such genetic changes have been discovered. Often these changes will be in a gene newly thought to be linked to developmental disorders and there will be little, if any, published literature to draw on. We then have to speculate whether the genetic change detected is the cause of our patient’s health problems, and whether any additional difficulties that have happened for our patient that have not yet been noted in other patients with changes in the same gene are an extension of the phenotype of the newly described disease gene, or coincidental. In situations like this, we are often unable to give people information about what a new diagnosis might mean for them or their child in the longer term.
This has led to patient support and awareness groups taking on an increasingly important role [ 16 ], as families gather to share their lived experience of newly diagnosed rare genetic conditions, in turn informing clinical services. For example, the charity Unique works with families and professionals to develop specialist information relating to many rare and newly described genetic conditions, and to gather information about their long-term effects, increasing awareness and understanding of what it is like to live with rare genetic conditions. The rapidity with which such information can be gathered is also exemplified by the work of the PURA Syndrome Foundation: in 2014 the first patients with a rare condition called PURA syndrome were described in the medical literature [ 17 ]. Shortly afterwards the PURA Syndrome Foundation was established which has catalysed links between families, clinicians and researchers, greatly improving the speed and quality of research into the condition [ 18 ].
The agnostic approach of exome and genome sequencing is also challenging our previous concepts of existing genetic diagnoses, when apparently pathogenic variants are found in well-described disease genes but the patient’s clinical picture falls outside the boundaries of what we would conventionally expect for a patient affected by that particular genetic condition. For example, loss-of-function variants in SOX2 are known to cause anophthalmia and microphthalmia in addition to other phenotypes such as developmental delay and structural brain anomalies. Eye abnormalities were thought to be a key feature of SOX2 -related disorders, and so SOX2 would only be requested as a genetic test in patients who had absent or small eyes. Recently, via ‘genotype-first’ approaches, loss-of-function SOX2 variants have been found in people with developmental delay but without anophthalmia or microphthalmia, broadening the phenotypic spectrum associated with this gene [ 19 ]. Case Study 1 shows a further example where exome testing has extended previous perceptions of the clinical scope of a genetic condition.
Case Study 1
Redefining our understanding of genetic conditions (fictional case based on eggens et al. [ 20 ]).
An 8-year-old girl was referred to clinical genetics in order to investigate her progressive weakness. She had been floppy as a baby and from the age of 5 years had developed worsening limb weakness with frequent unusual movements, and difficulty in swallowing. Serial brain scans had shown progressive cerebellar atrophy.
Exome testing found that she was homozygous for a variant predicted to disrupt the function of EXOSC3 , a gene associated with pontocerebellar hypoplasia. This diagnosis had never been thought of as she did not have one of the defining characteristics: pontine hypoplasia. Her clinical picture also seemed atypical for this condition – most children with pontocerebellar hypoplasia do not survive infancy.
However, recent research has shown genotype–phenotype correlations in EXOSC3 -mediated pontocerebellar hypoplasia – patients homozygous for p.D132A variants (like this patient had) tend to have a milder clinical course and preservation of the pons. This genetic explanation fitted well in retrospect, but would not have been considered in advance of the exome test.
Key messages
- Many well-recognised genetic conditions may have a wider spectrum of effects than previously thought.
- Patients with genetic conditions identified via genomic tests may not conform to the pattern we expect based on experience of patients with the same condition identified via single gene testing. It can be very difficult to be sure whether this reflects an incorrect diagnosis, or a wider disease spectrum than previously recognised.
In many cases, our understanding of why the same genetic condition may be expressed so differently among different people is at an early stage, and this often makes genetic counselling very challenging, particularly in the prenatal setting. For some genetic conditions, it is becoming possible to provide more personalised risk estimates, based on combining knowledge of a person’s genetic diagnosis, with analysis of other factors that may influence their risk. Personalisation of risk in this way has generally been crude and reliant on clinically obvious characteristics: for example, men with pathogenic BRCA variants have a lower risk of developing breast cancer than women with pathogenic BRCA variants. More recently, genetic testing is being developed to complement ‘key’ genetic test results to provide an increasingly refined personal risk. For example, use of a polygenic risk score using breast cancer and ovarian cancer susceptibility SNPs identified via population GWAS showed large differences in absolute cancer risks between women with pathogenic BRCA variants with higher compared with lower polygenic risk score values [ 21 ]. This has yet to translate into routine clinical practice, but has the potential to help women with pathogenic BRCA variants make more informed decisions about how and when to manage their cancer risk.
The downsides of improved sensitivity: increased uncertainty in what tests mean
The prior probability of any one variant identified via genome sequencing being causative for a patient’s rare disease is extremely low. Attempts to catalogue human genetic variation, for example via the 1000 Genomes Project, show that a typical human genome differs from the reference human genome at 4.1–5 million sites [ 22 ]. Most of these variations will be entirely benign, some may subtly impact on risk of various common diseases, and a very small number will have the potential to cause serious disease either in an individual, or in their children (potentially in combination with variants inherited from their partner).
Genome sequencing identifies the majority of these variants, which then need careful filtering to produce a meaningful output. This has required a significant change in mindset from an era when most variants were identified in the context of carefully chosen single gene sequencing, and so had a much higher prior probability of being causative. There is an increasing shift towards a view that variants should be ‘innocent until proven guilty’ [ 23 ], but there is a lack of consensus regarding how to translate this principle into clinical practice.
There is also considerable discrepancy in how different genetics laboratories interpret the same variants. International guidelines for variant interpretation are helpful but insufficient to remove a great deal of noise when attempting to assign significance to particular findings [ 24 ]. This was illustrated in a recent study comparing variant classification among nine genetic laboratories: although they all used the same guidelines, only 34% of variants were given the same classification by all laboratories, and 22% of variants were classified so differently that different medical interventions would be recommended [ 25 ]. At a lower resolution level, even being sure of the relationship between genes and diseases is often difficult. For example, curation of the 21 genes routinely available on Brugada syndrome gene panels using the ClinGen gene curation scoring matrix found that only one of these genes was definitively linked to Brugada syndrome [ 26 ]. Our improving knowledge of variant interpretation leaves us with a difficult legacy, with many patients having been diagnosed incorrectly with genetic conditions. The effects of this can be far-reaching and difficult to undo, as illustrated by Case Study 2 .
Case Study 2
The legacy of incorrect diagnosis (case reported by ackerman et al. [ 27 ]).
A teenage boy died suddenly and genetic testing was then undertaken for his brother, resulting in the finding of a rare variant in KCNQ1 . On the basis of this test, the living brother was diagnosed with long QT syndrome, and the teenage boy’s sudden death was attributed to long QT syndrome. The living brother had an implantable cardioverter defibrillator inserted, and via cascade genetic testing over 24 relatives were diagnosed as having long QT syndrome, despite having normal QT intervals on ECG.
However, subsequent examination of post-mortem samples found that the boy who died had cardiac features inconsistent with long QT syndrome, did not have the KCNQ1 variant found in the wider family, and instead had a clearly disease-causing de novo variant in DES , a gene linked to cardiomyopathy.
- It is very important to consider whether the clinical picture fits when evaluating variant significance: genetic variants will usually only predict disease well if found in the context of a medical or family history of the relevant disease.
- Incorrect (or inappropriately deterministic) genetic test interpretation can affect the clinical care of a whole family, not just the person being tested.
Although this suggests that we need to be very cautious in making firm genetic diagnoses, it is difficult to know where the threshold should lie for communicating genetic variation of uncertain significance. There is some evidence that people find receiving a variant of uncertain significance surprising and disturbing, and some people misinterpret it as being definitely pathogenic or definitely benign [ 28 ]. However, there is also evidence that many people have a strong desire to receive a broad range of results from genetic testing, including uncertain results, and are uncomfortable with the idea that decisions about non-disclosure might be made without involving them [ 29 ].
The fear is that disclosure of uncertain variants will lead to over-diagnosis and over-management, with variants inappropriately being treated as if pathogenic. Excessive and inappropriate interventions (not to mention anxiety and distress) might then cascade through families, going against one of the fundamental principles of medicine to ‘first do no harm’. However, we also fear missing something or being accused of ‘hiding information’. The result is that we tend to end up in purgatory, documenting uncertain variants on lab reports (though sometimes not) and having lengthy conservations with patients about them (though sometimes not), then tacking on a caveat that ‘maybe this means nothing’. This nominally shifts the responsibility to the next person in the chain but feels unsatisfactory for all concerned.
Uncertainty when to stop looking and what to communicate
Another issue arising from improved sensitivity is the ability to find genetic variants that are unrelated to the clinical problem that a patient presents with, but that may be relevant for their health in other ways. This may be viewed as positive or negative, but working out how to handle this information raises difficult questions. In 2013, the American College of Medical Genetics and Genomics (ACMG) suggested that laboratories should automatically seek and report pathogenic variants in 56 genes associated with ‘medically actionable’ conditions when performing clinical sequencing [ 30 ]. The main rationale was the potential to benefit patients and families by diagnosing disorders where preventative measures and/or treatments were available, with the aim of improving health. However, these recommendations proved controversial. The main debate at the time centred around whether patients should have a right to choose not to know such information [ 31 ]. Subsequent questions about the role of clinicians in offering additional findings, what constitutes a ‘medically actionable’ finding, and what is the predictive value of such findings in the absence of a phenotype or family history of the relevant disorder, are yet to be fully addressed.
Analysis of data from the 1000 Genomes cohort demonstrated that approximately 1% of ‘healthy’ people will have a ‘medically actionable’ finding in one of the 56 genes [ 32 ]. However, what this might mean on an individual basis is often unclear. Most of our knowledge regarding the effects of variation in any given gene has been gathered by observing people who have been identified as having variants in the gene because they were tested as they had a personal history or family history of disease, biasing the sample from which our conclusions are drawn. It is less clear what it might mean to find, for example, an apparently pathogenic variant in a gene linked to cardiomyopathy in a person with no personal or family history of heart problems. This has important implications for ‘cascade screening’, where relatives of a patient affected by a condition with a known genetic cause are offered testing to see whether they have the disease-causing genetic variant that was found in their clinically affected family member (meaning that they may also be at risk of developing the disease). To what extent should testing and subsequent screening be offered in a family based on an incidental finding of a genetic variant thought to be predictive of a particular condition, if there is no clinical evidence that anyone in the family, including the person in whom the genetic variant in question was first identified, is actually affected by it?
Broad genomic testing also has the potential to detect carrier status for recessive and X-linked conditions. From population studies, we know that being a carrier for a genetic condition is very common. For example, a gene panel testing carrier status for 108 recessive disorders in 23453 people found that 24% were carriers for at least one of the 108 disorders, and 5.2% were carriers for multiple disorders [ 33 ]. On a disorder-by-disorder status, being a carrier for a genetic condition is very rare (with notable exceptions such as haemochromatosis and cystic fibrosis), but when considered collectively, it is ‘normal’ to be a carrier for a genetic condition. For most people, being a carrier will have no impact on their life at all. However, if their partner happens to be a carrier for the same condition then the implications could be very profound, as each of their children would have a one in four chance of being affected by the genetic condition. This is particularly relevant for couples who are known to be biologically related [ 34 ], and couples with common ancestry, as they will have a higher chance of both being carriers for the same recessive condition. Carrier screening for various autosomal recessive diseases has been available in some instances for many years, for example screening for carrier status for Tay–Sachs disease for people of Ashkenazi Jewish ancestry has been offered since the 1970s [ 35 , 36 ]. More recently, advances in technology have led to development of expanded carrier screening tests, which check carrier status for multiple diseases simultaneously and are often less targeted towards particular genetic populations [ 37 ].
The increased scope of carrier screening, combined with the recognition that it is very common to be a carrier for one or more recessive genetic conditions, has led to an increasing move to consider carrier results for recessive genetic conditions on a couple basis, where carrier status is only communicated if it would be relevant in the context of a particular relationship (i.e. if both people in a couple are carriers for the same condition) [ 38 ]. This avoids pathologising the status of ‘being a carrier’, recognising that most of us are carriers for some genetic conditions, and conserves resources for genetics services by not flooding the system with large volumes of individual carrier results, most of which will be meaningless in the context of that individual’s life. Objections to this approach are that by not communicating individual carrier results, a person would not know this information for future relationships, and their family could not access cascade screening to see whether they are also carriers. However, these objections could be obviated by widespread adoption of couple carrier testing – a person (or their close relatives) could find out their carrier status if relevant when they next had a couple carrier test in the context of their new relationship. In some ways, this could be seen as comparable with management of infectious disease – lots of healthy people carry MRSA, but very few die of MRSA infection. People are therefore screened at times when they might be especially vulnerable to becoming unwell from MRSA, or when they might pass it on to others at risk, for example when admitted to hospital, rather than being tested at random points when they are generally well.
The expanding remit and availability of genetic technology
‘acute genetics’.
For many years, clinical genetics input has at times influenced acute care, for example in diagnosing trisomies in the neonatal period, or informing the care of babies born with ambiguous genitalia. However in many circumstances, the key contribution of clinical genetics was in providing a post hoc explanation for serious medical problems, rather than in influencing treatment decisions on a real-time basis. This is changing as the availability of exome and genome sequencing increases, as shown by Case Study 3 . A recent study in a neonatal intensive care unit in Texas studied outcomes for 278 infants who were referred for clinical exome sequencing, and found that 36.7% received a genetic diagnosis, and medical management was affected for 52% of infants with diagnoses [ 39 ]. There is increasing evidence that this approach is cost-effective: for example, a prospective study of exome sequencing for infants with suspected monogenic disorders found that standard care achieved an average cost per diagnosis of AU$ 27050, compared with AU$ 5047 for early singleton exome sequencing [ 40 ]. Similarly, ‘real-time’ genetic and genomic testing is making an impact in cancer treatment, where in many cases testing is available to help guide treatment choices by identifying actionable genetic variants in tumours that may respond to specific therapies [ 41 , 42 ].
Case Study 3
Insights from exome testing transforming a clinical course (case from wessex genomic medicine centre [ 43 ]).
A young woman was referred for exome testing having spent months in a coma. From childhood she had experienced sensory problems, and as a young adult she had gone on to develop seizures which deteriorated into status epilepticus, necessitating ventilation on intensive care.
After 3 years during which all other avenues had been explored, analysis of her exome was proposed. An unexpected diagnosis of pyridoxine-dependent epilepsy was found; this had not previously been considered as classically it causes seizures in the first few months of life. She began treatment with pyridoxine (vitamin B 6 ). From that point on she had no further seizures and her clinical situation transformed. Over a 6-month period she was weaned off all of her anti-epileptic drugs, and was able to return to a normal life.
Key message
- Exome or genome tests have the potential to make an enormous difference to clinical care and to people’s lives.
Pharmacogenomics
As well as guiding treatment choice, genetic testing will increasingly influence what doses are prescribed, and whether medications are considered unsuitable in view of a high risk of an adverse reaction. Around the time that the Human Genome Project was completed, there was considerable excitement about the possibility of genetic testing guiding use of medication in the clinic [ 44 , 45 ]. The potential of genotype-driven drug dosing has for the most part yet to be realised, in part because the interaction of the genetic factors involved is sometimes complex, and in part because environmental factors may also have a significant impact on how a person responds to a drug. For example, genotype-driven prescription of warfarin, which has notoriously wide inter-individual variation in dosage requirements, largely remains in the realm of research [ 46 ].
However, for some drugs, pharmacogenomics has already had a significant impact in reducing morbidity and mortality. For example, when the antiretroviral drug abacavir was first introduced, approximately 5% of the people treated developed an idiosyncratic hypersensitivity reaction that could be life-threatening on repeated exposure to the drug [ 47 , 48 ]. Research established that immunologically confirmed hypersensitivity reactions to abacavir only occurred in people with the HLA-B*5701 allele, and a clinical trial went on to show that pre-screening patients to check that they did not have HLA-B*5701 prior to starting the drug led to no confirmed hypersensitivity reactions in the pre-screened arm, while 2.4% of the unscreened patients had reactions [ 49 ]. Patients are now screened for HLA-B*5701 as standard before starting abacavir treatment [ 50 ]. Similar screening is likely to become more widespread as we learn more about genetic risk factors for adverse drug reactions. For example, there are increasing suggestions that the mitochondrial variant m.1555A>G should be checked in patients with cystic fibrosis in order to guide antibiotic treatment choices, in view of the evidence that people with this variant may develop hearing loss when exposed to aminoglycosides [ 51 ].
Evolving options in prenatal genetics
Genetic testing is also being used more extensively in the prenatal setting, in part because of developments in non-invasive prenatal testing and diagnosis, which allow genetic screening or testing of a developing pregnancy by doing a blood test for the mother [ 52 ]. This removes the risk of miscarriage associated with conventional prenatal tests (chorionic villus sampling or amniocentesis). While this is in some ways a stride forward, it raises various ethical issues, as the technical test safety may lead to such testing becoming viewed as routine. This raises the concern that couples will give less careful consideration as to whether they really want to know the results before having such tests, and that women may feel that there is an expectation that they should have testing. The worry is that this could potentially lead to people feeling under pressure to terminate pregnancies in response to genetic test results (including in situations where the clinical implications of the results may be far from clear) [ 53 ].
Widening access to genetic testing within healthcare
The expanding options for genetic testing and the escalating expectation for quick results to drive clinical management mean that testing provision is increasingly being pushed out of highly specialised genetics centres into mainstream medicine. For example, many women with ovarian cancer will now be offered BRCA testing via their oncology team, and only referred to genetics if needed based on the test results [ 54 ]. Genetics appointments now frequently focus on interpretation of tests already done, working out if the test outcome seems to match the clinical problem, and arranging testing and surveillance for family members.
The rise of direct-to-consumer genetic testing
As clinical services have increasingly grown to expect and demand genetic answers for patients with complex health problems, on a broader societal level the hunger for genetic information also seems to be increasing. However this is occurring in the context of a public discourse about personalised/precision medicine and genetics that tend to enthusiastically promote it in a very optimistic light, rarely dwelling on potential concerns and limitations, and therefore potentially sculpting inappropriate expectations from technology that is still being developed [ 55 ].
Direct-to-consumer tests currently sit outside much of the regulation that governs clinical genetic testing, but claim to provide insight into issues as diverse as ancestry, nutrition, athletic ability, and child talent [ 56 ]. Many testing providers also claim to help provide insight on health, though the information provided by many direct-to-consumer companies is far from comprehensive. For example, a recent analysis of 15 direct-to-consumer genetic testing companies advertising to U.K. consumers found that none of them complied with all the U.K. Human Genetics Commission principles for good practice regarding consumer information [ 57 ]. There are also examples that might make us reflect sceptically on the value of these tests – for example a case where a family sent a sample from their dog to a direct-to-consumer testing company designed to provide insights on people’s genetic ‘superpowers’ and received a report which did not mention that the sample was not human but conjectured that the client would be talented at basketball [ 58 ].
‘DIY genetics’ has also risen in popularity, with people asking for raw data from direct-to-consumer companies then processing this themselves via third-party interpretation services, as discussed in Case Study 4 . Approximately 40% of genetic changes in direct-to-consumer test raw data sent for clinical confirmation are false positives [ 59 ], but this is often not appreciated by customers or the doctors they may subsequently visit, leading to anxiety and often inappropriate medical interventions [ 60 ]. However, clearly many people see a value in receiving genetic information and are prepared to pay for this. This marks a shift from genetic testing in order to explain health problems or for people at high risk of developing specific genetic conditions, to testing of healthy people with the rationale of facilitating life planning. This idea has been taken to the extreme with initiatives such as the BabySeq project, exploring the medical, behavioural and economic impacts of integrating genome sequencing into the care of healthy newborns [ 61 ].
Case Study 4
Grime on the crystal ball (fictional case based on moscarello et al. [ 60 ]).
A healthy medical student was given a direct-to-consumer genetic test for Christmas, and explored the raw data from this test using an online interpretation programme, finding a variant in MYBPC3 that was predicted to cause hypertrophic cardiomyopathy. He was understandably worried by this result, taking time off university as he came to terms with it, and giving up running, which he used to really enjoy.
He was seen in a hypertrophic cardiomyopathy clinic and had an expert cardiology assessment including ECG, echocardiogram and review of his family history. He was found to have no clinical evidence of hypertrophic cardiomyopathy, and further genetic testing showed that he did not actually have the disease-causing MYBPC3 variant that the online interpretation programme had identified. However, he continued to feel anxious about his risk of heart problems and decided to give up running permanently.
- Information provided from direct-to-consumer testing may be unreliable, especially where online interpretation programmes are used to further explore the raw data from the test: the level of quality control may be very different from that of accredited genetic laboratories, increasing the likelihood of false positives, false negatives and sample mix-up.
- Many direct-to-consumer genetic tests involve no meaningful pre-test counselling – people are often totally unprepared for the information that might come out of such testing (and are unaware that it might be wrong).
Genetic information as family information
The familial nature of genetic information has always generated discussion as to how to respect the confidentiality of individual patients while ensuring that their close relatives have access to information that may be relevant for their own health and life choices. Clinical guidance in this area has increasingly taken the stance that genetic information should be confidential to families, not individuals (though the personal consequences of having a genetic change for a given individual should be confidential to them alone) [ 62 ].
The consequences of this shift are still being navigated in the clinical setting – research indicates that patients often see genetic information as belonging to their family rather than exclusively to them [ 63 ], but healthcare professionals are often reticent about taking a familial approach to the confidentiality of genetic information in practice, worrying that this stance could disrupt family dynamics or erode patient trust in the health service [ 64 ]. A recent BMJ poll which asked, ‘Are there situations when sharing a patient’s genetic information with relatives without consent is acceptable?’ demonstrated the current split in opinion, with 51% of respondents answering ‘yes’ and 49% ‘no’ [ 65 ]. The personal versus familial nature of genetic information is currently being tested in the courts via the ABC case, which centres around non-disclosure of genetic risk to the daughter of a patient with Huntington’s disease [ 66 ].
Treatment for genetic disorders
One of the most exciting recent developments in genetics and genomics is the prospect of treatment for an increasing number of genetic conditions. However this topic has to be treated with caution as the practical reality for many patients and families is that though promising research is ongoing, meaningful treatment is not possible in many cases. Even in situations where evidence-based treatments have been developed, the expense of many of these therapies risks making them inaccessible.
Many different approaches have been taken to try to treat genetic conditions. Gene therapy, which involves delivering functional genetic code, is one approach but its success has been widely variable, often due to difficulty in developing vectors that can deliver genetic material into affected tissues at sufficiently high levels without being destroyed by the immune system. In certain situations this approach can be highly effective, for example promising results have been achieved in various eye conditions, likely because eyes are small and easily accessible, and have a privileged relationship with the immune system [ 67 ]. In cases aiming to deliver gene therapy to a wider area, such as the lungs or the muscles, treatment attempts have generally proved more challenging [ 68 , 69 ].
Other approaches include use of small molecules to modify various steps in the pathway from gene to functional product. For example, Eteplirsen aims to treat Duchenne muscular dystrophy in certain patients by influencing splicing machinery to skip exon 51 from mature DMD mRNA, restoring a more functional reading frame so that a shortened version of dystrophin can be successfully translated [ 70 ]. Ivacaftor potentiates the action of CFTR channels in some patients with cystic fibrosis (G551D pathogenic variant) [ 71 ]. Enzyme replacement therapy is being trialled to treat children with mucopolysaccharidoses, for example idursulphase infusions in mucopolysaccharidosis type 2 [ 72 ].
While lots of these therapies are very exciting and show demonstrable changes at the molecular level in clinical trials, these cellular changes do not always clearly translate into improvements in clinically relevant outcomes. The therapies are also often hugely expensive, which raises very difficult ethical questions regarding whether limited resources should be spent on such treatments where there is often only limited proof of clinical efficacy.
However the increasing possibility of future treatments for genetic conditions is influencing clinical decisions around the care of very ill children. For example, recently nusinersen has shown promise as a treatment for some children with spinal muscular atrophy, but this may begin to raise new questions about whether interventions such as intubation and tracheostomy should be offered to infants with severe spinal muscular atrophy, where previously these would have been considered medically inappropriate [ 73 ]. This has consequences for the clinical conversations happening when these diagnoses are made. In the past, breaking news of such a diagnosis might flow naturally into discussions around palliation. The possibility of treatment now creates new options to consider, but also new challenges in considering with parents how best to care for their child [ 74 ]. The clinical impact and accessibility of emerging treatments is often very uncertain, but parents may prefer to explore even extremely long-shot treatments over accepting a palliative care pathway route, and may expect or seek crowd funding for experimental treatments for which there is as yet very little, if any, evidence of benefit.
Improving genetic technology has also had a significant impact on fertility services, ranging from pre-implantation genetic diagnosis to mitochondrial donation, offering new options for families affected by genetic conditions [ 75 , 76 ]. Increasing technological capability is set to extend the theoretically possible range of options – for example last year a group in China used the CRISPR/Cas9 system to correct pathogenic variants in the HBB and G6PD genes in human zygotes [ 77 ], though the efficiency and accuracy of the correction procedure was variable. This emerging possibility raises significant ethical issues which need debate. A recent report of the Nuffield Council on Bioethics on genome editing in the context of human reproduction suggested that there may be certain contexts in which this may be ethically acceptable, provided that such interventions were intended to secure the welfare of a person who may be born as a result, and that any such interventions would uphold principles of social justice and solidarity [ 78 ].
Conclusions
Insights from genomic technology have great potential to improve health, but we are currently going through a teething process in learning how to respond to the nebulous information that genomic tests can provide in the clinical setting. In part, this learning process is being driven by patients and families, with patient support groups coming to the fore in an era where we can now make extremely rare diagnoses that link different families across the world, but often have very little information on what this might mean for the future. Our current response to the outcomes from genomic tests is often reactive and ad hoc, partly because we are still learning how to interpret genomic variation and are often unable to gain a consensus on whether genetic variants are clinically significant or not. This situation is exacerbated by the different routes in which genomic information is now accessible – rapid tests to establish diagnosis or plan treatment for patients are now a reality in the real-life clinical setting, but healthy people also have increasing access to commercial tests that claim to provide genetic information to improve health and life planning. This raises particular challenges in the context of a public discourse about genomics that tends to present it as far more predictive and certain than it actually is. Some of the most exciting recent developments in genomic medicine relate to potential future treatments and reproductive options for people and families affected by rare genetic conditions. However hurdles relating to treatment efficacy and optimal timing of treatment, mean that we need to keep these advances in perspective and consider how to research potential treatments responsibly, avoiding creating hype that undermines the ability of families to make a balanced decision whether or not to participate in this research. It is also important to consider financial sustainability, avoiding situations where useful new treatments are developed that remain inaccessible to the patients who need them on account of their cost. To summarise, the introduction of genomic testing is having a big impact on patient care, but raises various issues that need further study and debate in order to help us maximise the potential benefits of genomic medicine while minimising the possible harms.
Acknowledgments
We thank the patient in Case Study 3 for her help with the Case Study box and for sharing her story.
Abbreviations
1 Complex chromosome rearrangements, thought to occur due to single catastrophic events where chromosomes ‘shatter’ and are repaired by error-prone mechanisms.
2 Clusters of localised mutations.
This work was supported by funding from a Wellcome Trust collaborative award [grant number 208053/Z/17/Z (to A.L.)].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
- International edition
- Australia edition
- Europe edition

Scientist who gene-edited babies is back in lab and ‘proud’ of past work despite jailing
China’s He Jiankui, who used Crispr to edit genome, says he is working on genetic diseases and suggests human embryo gene editing will one day be accepted
A Chinese scientist who was imprisoned for his role in creating the world’s first genetically edited babies says he has returned to his laboratory to work on the treatment of Alzheimer’s and other genetic diseases.
In an interview with a Japanese newspaper, He Jiankui said he had resumed research on human embryo genome editing, despite the controversy over the ethics of artificially rewriting genes, which some critics predicted would lead to demand for “designer babies”.
“We will use discarded human embryos and comply with both domestic and international rules,” He told the Mainichi Shimbun, adding that he had no plans to produce more genome-edited babies. Previously, He had used a tool known as Crispr-Cas9 to rewrite DNA in embryos.
In 2019 a court in China sentenced He to three years in prison for violating medical regulations after he claimed the previous year that he had created genetically modified twin sisters, Lulu and Nana, before birth.
His experiments sent shockwaves through the medical and scientific world. He was widely condemned for having gone ahead with the risky, ethically contentious and medically unjustified procedure with inadequate consent from the families involved.
The court found that He had forged documents from an ethics review panel that were used to recruit couples for his research.
He said he had used a gene-editing procedure known as Crispr-Cas9 to rewrite the DNA in the sisters’ embryos – modifications he claimed would make the children immune to HIV .
He has continued to defend his work, despite widespread criticism, saying he was “proud” of having created Lulu and Nana. A third girl was born in 2019 as a result of similar experiments.
He told the Mainichi that he hoped to use genome editing in human embryos to develop treatments for rare genetic diseases such as Duchenne muscular dystrophy and familial Alzheimer’s disease, at three laboratories he has opened since his release from prison in 2022.
He said the three genome-edited children were “perfectly healthy and have no problems with their growth”, according to the newspaper, adding that the twins, now aged 5, were attending kindergarten.
“The results of analysing [the children’s] entire gene sequences show that there were no modifications to the genes other than for the medical objective, providing evidence that genome editing was safe,” he told the Mainichi. “I’m proud to have helped families who wanted healthy children.”
He told the Guardian in 2023 that he had acted “too quickly” by pressing ahead with the procedure, but stopped short of voicing regret or apologising.
In his interview with the Mainichi, he said society would “eventually accept” human embryo gene editing in the quest to find treatments for genetic diseases.
Most viewed

How Can GARD Help You?
GARD Information Specialists offer individualized help to connect you with information and resources. Our Information Specialists can help you:
- Find or understand information about a rare disease.
- Navigate information throughout the diagnostic journey.
- Discover resources, disease experts, or clinical studies for a rare disease.
Available Monday through Friday 12 pm to 6 pm Eastern Time (Except: Federal Holidays )
1-888-205-2311
Contact gard.
Use the contact form to send your question to a GARD Information Specialist. Please allow 2 to 10 business days for us to respond.
Connect with GARD

- Introduction to Genomics
- Educational Resources
- Policy Issues in Genomics
- The Human Genome Project
- Funding Opportunities
- Funded Programs & Projects
- Division and Program Directors
- Scientific Program Analysts
- Contact by Research Area
- News & Events
- Research Areas
- Research investigators
- Research Projects
- Clinical Research
- Data Tools & Resources
- Genomics & Medicine
- Family Health History
- For Patients & Families
- For Health Professionals
- Jobs at NHGRI
- Training at NHGRI
- Funding for Research Training
- Professional Development Programs
- NHGRI Culture
- Social Media
- Broadcast Media
- Image Gallery
- Press Resources
- Organization
- NHGRI Director
- Mission & Vision
- Policies & Guidance
- Institute Advisors
- Strategic Vision
- Leadership Initiatives
- Diversity, Equity, and Inclusion
- Partner with NHGRI
- Staff Search
Genetic Disorders
Many human diseases have a genetic component. Some of these conditions are under investigation by researchers at or associated with the National Human Genome Research Institute (NHGRI).
A genetic disorder is a disease caused in whole or in part by a change in the DNA sequence away from the normal sequence. Genetic disorders can be caused by a mutation in one gene (monogenic disorder), by mutations in multiple genes (multifactorial inheritance disorder), by a combination of gene mutations and environmental factors, or by damage to chromosomes (changes in the number or structure of entire chromosomes, the structures that carry genes). As we unlock the secrets of the human genome (the complete set of human genes), we are learning that nearly all diseases have a genetic component. Some diseases are caused by mutations that are inherited from the parents and are present in an individual at birth, like sickle cell disease. Other diseases are caused by acquired mutations in a gene or group of genes that occur during a person's life. Such mutations are not inherited from a parent, but occur either randomly or due to some environmental exposure (such as cigarette smoke). These include many cancers, as well as some forms of neurofibromatosis.
List of Genetic Disorders
This list of genetic, orphan and rare diseases is provided for informational purposes only and is by no means comprehensive.

Featured Content

Last updated: May 18, 2018
NIH VideoCasting
CIT can broadcast your seminar, conference or meeting live to a world-wide audience over the Internet as a real-time streaming video. The event can be recorded and made available for viewers to watch at their convenience as an on-demand video or a downloadable file. CIT can also broadcast NIH-only or HHS-only content.
VideoCast Send Live Feedback
Clinical Center Grand Rounds: Great Teachers Distinguished Clinical Research Scholar and Educator in Residence Lecture - Genetics & Genomics of Neuropsychiatric Disorders
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
April 2, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source

New study finds mental health emergencies in kids were more severe during the pandemic
by Ann & Robert H. Lurie Children's Hospital of Chicago

A new study has found that during the pandemic pediatric emergency departments (EDs) saw more children and adolescents who needed a psychiatric admission, as well as an increase in severe conditions, such as bipolar disorder, schizophrenia and substance use disorders.
The higher demand for a psychiatric inpatient bed often exceeded availability, resulting in over 12-hour stays in the ED awaiting admission for nearly 20% of children with mental health emergencies in 2022, up from 7% before the pandemic . Findings were published in Academic Emergency Medicine .
"Our data shows that pediatric emergency departments saw more severe mental health presentations during the pandemic, even while the actual number of visits decreased in 2022," said lead author Jennifer Hoffmann, MD, MS, emergency medicine physician at Ann & Robert H. Lurie Children's Hospital of Chicago and Assistant Professor of Pediatrics at Northwestern University Feinberg School of Medicine.
"The dramatic increase in prolonged ED stays attests to the strain on the system and difficulties finding appropriate psychiatric care for children, whether in the hospital or in the community."
Dr. Hoffmann and colleagues retrospectively studied mental health ED visits by children aged 5 to less than 18 years at nine U.S. hospitals participating in the Pediatric Emergency Care Applied Research Network Registry from 2017 to 2022. They described these visits by period—pre-pandemic (January 2017–February 2020), early pandemic (March 2020–December 2020), mid pandemic (2021) and late pandemic (2022).
In addition to the increased severity of mental health emergencies, they found that during the mid and late pandemic, mental health ED visits increased beyond expected rates among girls, but not among boys.
"We observed a unique vulnerability for girls during the pandemic, which indicates that girls' mental health requires more attention," said Dr. Hoffmann, who also is the Children's Research Fund Junior Board Research Scholar at Lurie Children's.
Explore further
Feedback to editors

New report presents a global plan to combat prostate cancer
2 hours ago

Dogs may provide new insights into human aging and cognition
AI's ability to detect tumor cells could be key to more accurate bone cancer prognoses
3 hours ago
Novel pre-clinical models help advance therapeutic development for antibiotic-resistant bacterial infections

Far-UVC light can virtually eliminate airborne virus in an occupied room, study shows
4 hours ago

Las Vegas mass shooting survivors continue to struggle with major depression, PTSD

Combining food taxes and subsidies can lead to healthier grocery purchases for low-income households
New insights into muscle health: How innervation influences the recovery process
5 hours ago
Simulations reveal mechanism behind protein buildup in Parkinson's disease

Increasing positive affect in adolescence could lead to improved health and well-being in adulthood
6 hours ago
Related Stories

Children's mental health visits to emergency departments increased during COVID-19 pandemic
Jun 28, 2022

Emergency departments saw firearm injuries in children double during pandemic
Nov 6, 2023

Long wait times increase for children seeking emergency care for mental health
Apr 5, 2021

More than 1 in 10 pediatric ambulance runs are for mental health emergencies, finds study
Nov 16, 2023

Nearly half of children on Medicaid lack outpatient follow-up within a month after emergency care for mental health
Feb 13, 2023

Study: Increases in pediatric mental health emergency visits persist throughout pandemic
Oct 20, 2023
Recommended for you

Exploring the factors that influence people's ability to detect lies online
10 hours ago

Infant gut microbes have their own circadian rhythm: Study finds diet has little impact on how the microbiome assembles
9 hours ago

Pilot study shows ketogenic diet improves severe mental illness
Apr 1, 2024
International study uses AI to show how personality influences the expression of our genes
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
Could the Keto Diet Help Ease Psychiatric Conditions?
By Ernie Mundell HealthDay Reporter

TUESDAY, April 2, 2024 (HealthDay News) -- Patients with schizophrenia or bipolar disorder tend to see their conditions ease after four months on the ketogenic ("keto") diet, a small pilot study finds.
While no one is saying the diet should replace standard medications, the researchers believe it could provide additional help for some.
“It’s very promising and very encouraging that you can take back control of your illness in some way, aside from the usual standard of care,” said study first author Dr. Shebani Sethi . She's an associate professor of psychiatry and behavioral sciences at Stanford University.
The findings were published March 27 in the journal Psychiatric Research .
U.S. Cities With the Most Homelessness

Sethi said she first noticed there might be a connection between the keto diet and psychiatric health when she was working as a student in a clinic focused on obesity and weight loss.
Many people with psychiatric conditions gain excess weight due to medication side effects. Sethi was helping to treat one such patient, who had schizophrenia.
The patient's auditory hallucinations ("hearing voices" can be a common symptom of schizophrenia) quieted down after being on the keto diet, she said.
A search of the literature turned up little regarding using the diet to counter schizophrenia, but there was evidence it could ease epileptic seizures.
Apparently the diet did so "by reducing the excitability of neurons in the brain,” Sethi explained in a Stanford news release. “We thought it would be worth exploring this treatment in psychiatric conditions.”
The new trial was small -- just 21 adults diagnosed with either schizophrenia or bipolar disorder . All were on medication for their conditions and also had excess weight gain, troubles with high cholesterol or blood sugar control and/or insulin resistance.
They were all told to follow the ketogenic diet -- in this case, a regimen that included about 10% of calories coming from carbohydrates, 30% from protein and 60% from fat.
“The focus of eating is on whole non-processed foods including protein and non-starchy vegetables, and not restricting fats,” explained Sethi. Participants received keto cookbooks and access to a health coach.
Blood tests were used to measure just how closely folks stuck to the regimen.
Weight loss was the first big plus: Participants lost an average of 10% of their body weight and 11% off their waistline measurements, the researchers reported.
Patients also benefited from reductions in blood pressure, triglycerides (a kind of blood fat), blood sugar levels and insulin resistance.
“We’re seeing huge changes,” Sethi said. “Even if you’re on antipsychotic drugs, we can still reverse the obesity, the metabolic syndrome, the insulin resistance. I think that’s very encouraging for patients.”
Patients' brains seemed to benefit, too.
“The participants reported improvements in their energy, sleep, mood and quality of life,” Sethi said. “They feel healthier and more hopeful.”
Mental benefits were tracked using what's know as the clinical global impressions scale. On average, folks on the keto diet saw a 31% improvement in scores, the Stanford team said.
Just how is a change in diet helping?
According to Sethi, evidence is gathering that illnesses such as schizophrenia and bipolar disorder may stem, in part, from metabolic deficits in the brain, which affect neuronal "excitability."
“Anything that improves metabolic health in general is probably going to improve brain health anyway,” Sethi said. “But the ketogenic diet can provide ketones as an alternative fuel to glucose for a brain with energy dysfunction.”
Most people were also able to stick to the keto diet, the study found. Fourteen of the 21 participants were "fully adherent," six were semi-adherent and only one was non-adherent.
More information
Find out more about the ketogenic diet at Harvard Health .
SOURCE: Stanford University, news release, April 1, 2024
Copyright © 2024 HealthDay . All rights reserved.
Join the Conversation
Tags: psychology , diet and nutrition
America 2024

Health News Bulletin
Stay informed on the latest news on health and COVID-19 from the editors at U.S. News & World Report.
Sign in to manage your newsletters »
Sign up to receive the latest updates from U.S News & World Report and our trusted partners and sponsors. By clicking submit, you are agreeing to our Terms and Conditions & Privacy Policy .
You May Also Like
The 10 worst presidents.
U.S. News Staff Feb. 23, 2024

Cartoons on President Donald Trump
Feb. 1, 2017, at 1:24 p.m.

Photos: Obama Behind the Scenes
April 8, 2022

Photos: Who Supports Joe Biden?
March 11, 2020

EXPLAINER: Rare Human Case of Bird Flu
Cecelia Smith-Schoenwalder April 2, 2024

The 7 Worst U.S. Presidential Scandals
Seth Cline March 1, 2017

Trump Hits the Trail Amid Legal Dramas
Lauren Camera April 2, 2024

Labor Market Keeps on Chugging
Tim Smart April 2, 2024

Florida High Court OKs Abortion Vote
Lauren Camera April 1, 2024

The Week in Cartoons April 1-5
April 2, 2024, at 12:32 p.m.

New research advances understanding of cancer risk in gene therapies
Medical research has shown promising results regarding the potential of gene therapy to cure genetic conditions such as sickle cell disease and the findings of this study, published in Nature Medicine , offer important new insights into processes happening in the body after treatment.
The present study looked at samples from six patients with sickle cell disease who were undergoing gene therapy as part of a major clinical trial at Boston Children's Hospital. The research brought together an international team of experts, to take a closer look at the genetic changes in the stem cells of patients before and after gene therapy and compare the cells that were modified with the therapy to those that weren't.
The study highlights the importance of long-term and in-depth monitoring of stem cell samples from patients with genetic conditions to track mutations that could lead to blood cancer, the researchers say.
Co-senior author of the study Professor David Kent from the Department of Biology and York Biomedical Research Institute said: "Think of the gene therapy process like clearing a forest and planting new seedlings. If you imagine that some seedlings are red and some are green, then the 'selection' we are observing is akin to having the forest regrow red trees preferentially.
"Our research indicates that gene therapy imposes a selection on different blood stem cells, the 'seed' cells of our blood and immune system. After gene therapy, the treatment might favour growth of stem cells with certain mutations, and this in turn could potentially lead to expansion of blood cells containing these mutations. In other settings, such expansions have been associated with development of blood cancers, particularly in older individuals, but the relationship of this study's findings to the risk of blood cancers is not yet fully understood."
The researchers used new technologies in genome science that allow blood cells to be tracked and compared in patients, a new approach which could substantially influence gene therapy trials in the future.
Co-senior author of the study Dr David Williams from Boston Children's Hospital and Harvard Medical School noted: "Gene therapy holds immense potential to cure genetic conditions such as sickle cell disease, and understanding how the process influences blood stem cell growth in the long run is crucial for safety. Notably, our study revealed that younger patients, with fewer genetic mutations in their stem cells, didn't exhibit strong signs of mutations post-therapy. This suggests that treating patients with gene therapy at a younger age could be both safer and more effective, but substantial work needs to be done to test this formally."
Sickle cell disease, an inherited genetic disorder, alters the natural round and flexible shape of red blood cells into a sickle or crescent shape, leading to severe health issues and putting patients at higher risk of developing blood cancer. Though approximately 100 million people carry the sickle cell trait worldwide, the disease only occurs if a child inherits the trait from both parents. In regions where the trait is prevalent, such as parts of sub-Saharan Africa, up to 20 percent of the population can be affected.
However, recent advances in gene therapy offer hope. This innovative approach involves modifying a patient's own stem cells outside the body, correcting the faulty gene responsible for the abnormal cell shape. These corrected cells are reintroduced into the patient, aiming to replace the problematic cells with healthy, normal-shaped ones.
The study, funded by the Bill and Melinda Gates Foundation, reveals that the gene therapy treatment itself is not the likely cause of new DNA mutations in blood stem cells. Instead, the process of genetically modifying these stem cells outside the body and re-transplanting them back into the patient makes blood stem cells that already have these mutations more prominent, thereby increasing their influence on the blood and immune systems.
The research team say the findings also suggest that younger patients may have acquired fewer stem cell mutations in their lifetime, which may inform the optimal age for gene therapies in this and other diseases in the future.
Co-lead author of the paper, Dr Alyssa Cull from the Department of Biology and York Biomedical Research Institute, emphasised the need for further research: "We now require more in-depth studies to uncover the precise connections behind specific mutations and the gene therapy procedure. There are pressing questions regarding how we can refine gene therapy to avoid stem cells that might contain mutations that affect blood cell growth. Determining whether mutations within a sample of cells are dangerous in the long run remains challenging though and substantial further research is needed."
Co-lead author Michael Spencer-Chapman from the Wellcome Sanger Institute echoes the need for longer term follow-up of patients saying: "Our research revealed that both gene-therapy modified and unaltered stem cells from sickle cell patients sometimes contained cancer-associated mutations leading to accelerated growth through the gene therapy procedure. Continuously tracking these mutations and gaining a deeper understanding of these processes will profoundly impact the future well-being of sickle cell patients around the world."
- Gene Therapy
- Sickle Cell Anemia
- Immune System
- Personalized Medicine
- Medical Topics
- Diseases and Conditions
- Embryonic stem cell
- Gene therapy
- Adult stem cell
- Alzheimer's disease
- Stem cell treatments
Story Source:
Materials provided by University of York . Note: Content may be edited for style and length.
Journal Reference :
- Michael Spencer Chapman, Alyssa H. Cull, Marioara F. Ciuculescu, Erica B. Esrick, Emily Mitchell, Hyunchul Jung, Laura O’Neill, Kirsty Roberts, Margarete A. Fabre, Nicholas Williams, Jyoti Nangalia, Joanne Quinton, James M. Fox, Danilo Pellin, Julie Makani, Myriam Armant, David A. Williams, Peter J. Campbell, David G. Kent. Clonal selection of hematopoietic stem cells after gene therapy for sickle cell disease . Nature Medicine , 2023; DOI: 10.1038/s41591-023-02636-6
Cite This Page :
Explore More
- Speed of Visual Perception Ranges Widely
- 3D Printed Replica of an Adult Human Ear
- Extremely Fast Wound Healing: New Treatment
- Micro-Lisa! Novel Nano-Scale Laser Writing
- Simple Brain-Computer Link: Gaming With Thoughts
- Clinical Reasoning: Chatbot Vs Physicians
- Understanding People Who Can't Visualize
- Illuminating Oxygen's Journey in the Brain
- DNA Study IDs Descendants of George Washington
- Heart Disease Risk: More Than One Drink a Day
Trending Topics
Strange & offbeat.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Clinical genetics articles from across Nature Portfolio
Clinical genetics involves the study, counselling and treatment of individuals and families with heritable disorders and disease predisposition. Diagnostic tools include standard ontologies for describing dysmorphology and traits, pedigree analysis, disease locus mapping by linkage or homozygosity, karyotyping, genome sequencing and genotyping.
Workshop report: the clinical application of data from multiplex assays of variant effect (MAVEs), 12 July 2023
Clinical classification of genomic variants identified on sequencing is often challenging, with many variants classified as Variants of Uncertain Significance (VUS) on account of insufficient evidence. Advances in sequencing and gene synthesis has made feasible multiplexed assays of variant effect (MAVEs), which quantify the functional impact of many thousands of genomic variants in a single experiment. These assays and the functional evidence they generate have the potential to empower more accurate clinical variant classification. However, there are many outstanding challenges and opportunities that require joint resolution and specification, thus necessitating communication between the research scientists who have designed and performed MAVEs and the clinicians and diagnostic scientists who will apply their data to clinical variant classification. In the ‘Clinical Application of MAVE Data’ workshop, held on 12th July 2023 at the Wellcome Connecting Science Conference Centre in between two relevant research meetings, ‘Curating the Clinical Genome 2023’ and the ‘Mutational Scanning Symposium 2023’, 44 key scientific and/or clinical stakeholders were brought together to consider important questions relating to clinical application of MAVE data, such as quantitative validation, variant truth-sets, platforms and standards for dissemination of MAVE data. The outcomes and possible next steps that were discussed encompassed development of focused workshops to develop consensus recommendations, creating a MAVE evaluation working group, and collaboration of ClinVar and MaveDB to enact software changes that support enhanced functional data submission.
- Sophie Allen
- Alice Garrett
- Clare Turnbull
Related Subjects
- Cancer genetics
- Chromosome abnormality
- Clinical epigenetics
- Disease genetics
- Gene therapy
- Genetic testing
Latest Research and Reviews
De novo variants in GABRA4 are associated with a neurological phenotype including developmental delay, behavioral abnormalities and epilepsy
- Samin A. Sajan
- Ralph Gradisch
- Martin Krenn
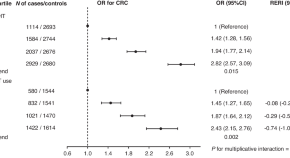
Genetic risk impacts the association of menopausal hormone therapy with colorectal cancer risk
- Jenny Chang-Claude
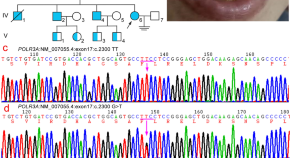
Clinical phenotype and genetic function analysis of a family with hypomyelinating leukodystrophy-7 caused by POLR3A mutation
- Dan-dan Ruan
- Xing-lin Ruan
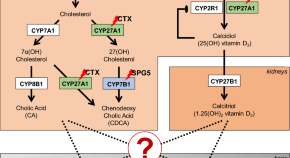
Vitamin D 3 deficiency and osteopenia in spastic paraplegia type 5 indicate impaired bone homeostasis
- Sabrina Ehnert
- Stefan Hauser
- Tim W. Rattay
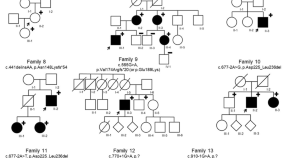
Expanding the clinical spectrum of biglycan-related Meester-Loeys syndrome
- Josephina A. N. Meester
- Anne Hebert
- Bart L. Loeys

Secondary use of genomic data: patients’ decisions at point of testing and perspectives to inform international data sharing
- Melissa Martyn
- Emily Forbes
News and Comment
Advancing access to genome sequencing for rare genetic disorders: recent progress and call to action.
- Vaidehi Jobanputra
- Brock Schroeder
- Ryan J. Taft
Stealthy stem cells to treat disease
Gene-editing strategies that allow stem cells to evade the immune system offer hope for universal cell-replacement therapies.
- Elie Dolgin
‘Epigenetic’ editing cuts cholesterol in mice
Changes to chemical tags on DNA in mice dial down the activity of a gene without cuts to the genome.
- Heidi Ledford
Considerations for clinical implementation of polygenic risk scores in diverse US populations
This study seeks to highlight the scientific, regulatory and operational issues around the use of polygenic risk scores in a diverse population. The work presented here provides a framework for laboratories, providers and researchers wishing to advance the field of preventative medicine.
Gene-editing breakthrough for a rare hereditary disorder
Patients who received an in vivo CRISPR-based gene-editing therapy for hereditary angioedema showed no serious side effects and nearly complete disease control.
- Karen O’Leary
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

- Got a Scoop?
- Arts & Entertainment
- Food & Drink
- Real Estate
- Indy Parenting
- Cover Stories
- Classifieds
- Create Event
Genetic Research Aids in Naming New Rare Succulent Species from Orange County
Share this:.
- Click to share on Facebook (Opens in new window)
- Click to share on X (Opens in new window)
- Click to email a link to a friend (Opens in new window)
- Click to print (Opens in new window)
Press releases are posted on Independent.com as a free community service.
Santa Barbara, Calif. – March 27, 2024 – A new rare species of succulent plant from Orange County has been named by Stephen McCabe, emeritus director of research at the UC Santa Cruz Arboretum, Kristen Hasenstab-Lehman, Ph.D., conservation geneticist and lab manager at Santa Barbara Botanic Garden, and Matt Guilliams, Ph.D., Tucker Systematist and curator of the Clifton Smith Herbarium at Santa Barbara Botanic Garden. The naming of new species is part of an effort by multiple entities to help conserve the many threatened species in the genus Dudleya, which are also known, ironically, as liveforevers.
The species, Dudleya chasmophyta, the crevice-loving Dudleya , was named in the California Botanical Society journal Madroño. When naming the plant, McCabe, along with colleagues Hasenstab-Lehman and Guilliams, opted not to name it after a specific person or place. Instead, they derived its new name from its affinity for dwelling in crevices, thus the name chasmophyta. Local experts Fred Roberts and Ron Vanderhoff of the California Native Plant Society helped arrange access to the plants via the U.S. Forest Service, using their knowledge of Orange County botany.
Although the range of the new species is incredibly small, occupying one cliff band in one canyon, it is not as threatened by poaching as other species of Dudleya that have been in the news in the last few years. That’s because the cliffs on which it grows are steep and have friable rocks that make climbing unsafe. Private property, thorny shrubs, and poison oak also protect the plants. Two fires have impacted surrounding areas and the top and bottom of the population, but it is likely that most of the plants have survived the fires, according to Roberts and McCabe. Vanderhoff notes that this is the second species to be known only from Orange County, the other being the Laguna Beach Dudleya.
Dudleya chasmophyta had rarely been visited and was previously considered to be part of a federally endangered species from Malibu Canyon in Los Angeles County. David Verity found the original Orange County plants in about 1950 and considered them likely to be the same as the ones in Malibu Canyon 60+ miles away. Unfortunately, he did not preserve a specimen.
Genetic work conducted by Hasenstab-Lehman provided supporting evidence that the species was distinct. “ Our genomic data provided clear evidence that the new species, D. chasmophyta, did not share the same branch in the evolutionary tree of life with the federally threatened Santa Monica Mountains Dudleya (Dudleya cymosa ssp.ovatifolia), a subspecies of the Malibu Canyon Dudleya,” Hasenstab-Lehman explains. “We know now both these rare liveforevers are even more restricted in the areas they live on the California Coast.”
With the separation into two species and after fires in Malibu Canyon, the Malibu Canyon Dudleya is now considered even more critically rare than when it was listed as endangered. “ Without the name, many of the agencies will not recognize it as something that needs protecting, ” explains McCabe. Seeds have now been collected for seed banks in case poaching, climate change, landslides, or some catastrophe affects the new species.
The different gardens and groups concerned with the rare Dudleya are working to preserve habitat, seed-bank populations, conduct research, and educate the public about buying responsibly propagated plants, not plants poached from the wild.
Hasenstab-Lehman, Guilliams, and McCabe have submitted genetic research for publication that bolsters their argument for naming the new species. McCabe is also finishing up a phase of a long-term research project with Channel Islands National Park by publishing an article about Dudleya of the Channel Islands. The article in the Cactus and Succulent Journal may help the park, the Nature Conservancy, and the Navy plan for conservation of Dudleya on those islands.
Get News in Your Inbox
Please note this login is to submit events or press releases. Use this page here to login for your Independent subscription
Username or Email Address
Remember Me
Not a member? Sign up here.

IMAGES
VIDEO
COMMENTS
Scientists successfully treated a rare disease with the experimental gene-editing technique. It could open the door to new ways of treating more common disorders in the future.
RSS Feed. Genetics research is the scientific discipline concerned with the study of the role of genes in traits such as the development of disease. It has a key role in identifying potential ...
"We demonstrated that genotype-first research can work, especially for identifying people with rare disorders who otherwise might not have been brought to clinical attention," says Caralynn Wilczewski, Ph.D., a genetic counselor at the National Human Genome Research Institute's (NHGRI) Reverse Phenotyping Core and first author of the paper.
In 1987, the New York Times Magazine characterized the Human Genome Project as the "biggest, costliest, most provocative biomedical research project in history." 2 But in the years between the ...
Gene therapy techniques are aimed at treating a variety of human diseases, including dominant and recessive genetic conditions, cancer, cardiovascular disease, and neurodegenerative diseases.
RSS Feed. Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring ...
New approach successfully traces genomic variants back to genetic disorders. ScienceDaily . Retrieved March 31, 2024 from www.sciencedaily.com / releases / 2023 / 01 / 230105151305.htm
A new study has identified 17 genetic variants that may influence Alzheimer's disease risk, putting researchers one step closer to uncovering biological pathways to target for future treatment and ...
Fetuses make a protein that causes morning sickness in pregnancy. A hormone called GDF15 triggers a part of the brain involved in nausea and vomiting, a new study finds. Blocking its action may ...
"The study shows that 81% of patients have abnormal teeth, while 59% of patients have skin issues and 27% have nail issues. Approx. 1/3 of patients sweat less than normal, leading to a risk of ...
Scientists Extract Genetic Secrets from 4,000-Year-Old Teeth to Illuminate the Impact of Changing Human Diets Over the Centuries. Mar. 27, 2024 — Researchers have recovered remarkably preserved ...
Researchers at the National Institutes of Health have discovered a new genetic disorder characterized by developmental delays and malformations of the brain, heart and facial features. Named linkage-specific-deubiquitylation-deficiency-induced embryonic defects syndrome (LINKED), it is caused by a mutated version of the OTUD5 gene, which ...
April 10, 2023 • Research Highlight. Epilepsies are chronic neurological disorders in which large groups of neurons firing at the same time generate electrical activity that causes seizures and involuntary movements. They are one of the most common brain diseases in children and, in almost a quarter of cases, patients do not respond to standard medical treatments.
Participants in clinical studies help current and future generations. Through these studies, researchers develop new diagnostic tests, more effective treatments, and better ways of managing diseases with genetic components. Participants in studies are actively involved in understanding their disorder and current research.
Tweaking a gene can either increase or decrease growth, and this is made more complex considering these dials are interconnected with each other, like cogs in a machine. While scientists are now ...
Genetic rare diseases are the focus of our article, accounting for nearly 80% of all rare diseases (Boat, 2015, Wright et al., 2018). However, rare diseases also include non-genetic rare diseases such as autoimmune disorders and rare cancers as well as maladies caused by infectious or toxic agents (Orphanet, 2020). Some are known to the general ...
New disease gene discovery and changing concepts of diagnosis. Exome and genome sequencing are powerful diagnostic tools - for example the Deciphering Developmental Disorders project, which recruited patients with severe undiagnosed disorders (who had generally already had any currently available diagnostic genetic testing), achieved a 40% diagnosis rate via trio exome sequencing for the ...
Medical genetics articles from across Nature Portfolio. Medical genetics involves the application of genetics to medical care, including research on the causes and inheritance of genetic disorders ...
China's He Jiankui, who used Crispr to edit genome, says he is working on genetic diseases and suggests human embryo gene editing will one day be accepted A Chinese scientist who was imprisoned ...
1-888-205-2311. Contact GARD. Use the contact form to send your question to a GARD Information Specialist. Please allow 2 to 10 business days for us to respond. Ask a GARD Specialist. GARDGenetic and Rare Diseases. Information Center. PO Box 8126, Gaithersburg, MD 20898-8126.
A genetic disorder is a disease caused in whole or in part by a change in the DNA sequence away from the normal sequence. Genetic disorders can be caused by a mutation in one gene (monogenic disorder), by mutations in multiple genes (multifactorial inheritance disorder), by a combination of gene mutations and environmental factors, or by damage to chromosomes (changes in the number or ...
New genetic analysis tool tracks risks tied to CRISPR edits. ScienceDaily . Retrieved March 28, 2024 from www.sciencedaily.com / releases / 2024 / 03 / 240326170107.htm
Doctors are feeling unable to tackle the growing problem of childhood obesity due to a lack of training and capacity according to new research. In a paper published in the British Journal of ...
As this extended genetic region is known for its pathological association with many neurodegenerative disorders including Alzheimer's disease, we investigated whether the LIFO brain regions ...
<p>Genetics & Genomics of Neuropsychiatric Disorders</p><p><br></p><p>Daniel Geschwind MD PhD<br>Gordon and Virginia MacDonald Distinguished Professor of Human Genetics<br>Neurology and Psychiatry<br>Senior Associate Dean and Associate Vice Chancellor of Precision Health<br>Institute for Precision Health<br>University of California Los Angeles</p><br>For more information go to <a href ...
New study maps a group of rare genetic diseases for the first time 1 hour ago Early cortical remyelination has a neuroprotective effect in multiple sclerosis, study shows
US News is a recognized leader in college, grad school, hospital, mutual fund, and car rankings. Track elected officials, research health conditions, and find news you can use in politics ...
Medical research has shown promising results regarding the potential of gene therapy to cure genetic conditions such as sickle cell disease and the findings of this study, published in Nature ...
RSS Feed. Clinical genetics involves the study, counselling and treatment of individuals and families with heritable disorders and disease predisposition. Diagnostic tools include standard ...
Santa Barbara, Calif. - March 27, 2024 - A new rare species of succulent plant from Orange County has been named by Stephen McCabe, emeritus director of research at the UC Santa Cruz Arboretum, Kristen Hasenstab-Lehman, Ph.D., conservation geneticist and lab manager at Santa Barbara Botanic Garden, and Matt Guilliams, Ph.D., Tucker Systematist and curator of the Clifton Smith Herbarium at ...